Liver tissue microbiota in nonalcoholic liver disease: a change in the paradigm of host-bacterial interactions
Nonalcoholic fatty liver disease (NAFLD) pathogenesis is explained by the complex relationship among diet and lifestyle-predisposing factors, genetic variance of the nuclear and mitochondrial genome, special characteristics of the phenome, including obesity, and the yet not fully explored interactions with epigenetic and other environmental factors, including the microbiome (1).
Despite the wealth of knowledge gained from molecular and genome-wide investigations in patients with NAFLD, the precise mechanisms that explain the variability of the histological phenotypes are not fully understood (2,3). Earlier studies on the gut microbiota in patients with NAFLD and nonalcoholic steatohepatitis (NASH) provided clues on the feces bacterial profile/s associated with the disease (4-19) (Figure 1). Nevertheless, the composition of the gut microbiota does not fully explain tissue-specific mechanisms associated with the degree of disease severity, including liver inflammation, ballooning of hepatocytes, and fibrosis. Here, we highlight the evidence on the role of liver tissue bacterial DNA in the biology of NAFLD and NASH. Besides, we provide evidence of metagenomic findings that can serve as the seed of further hypothesis-raising studies as well as can be leveraged to discover novel therapeutic targets.
Non-intestinal microbiotas and liver diseases
With the explosion of knowledge in the field of metagenomics, novel concepts on the pathogenesis of complex diseases emerged, including the putative role of tissue microbiotas.
A remarkable seminal study that surveyed bacteria profiles from up to 27 different body sites from healthy adults over time found that bacterial composition is primarily determined by the body habitat (20). This study demonstrated that the human microbiota varies systematically across body habitats, and it has thus established the basis onto which to explore tissue microbiotas in different disease states (20).
Explorations from studies in the past decade, particularly in the field of cardiovascular disease, suggested that the microbial composition of non-intestinal tissues may contribute to the pathogenesis of non-transmissible complex diseases. For example, Koren et al. used pyrosequencing of 16S rRNA genes to survey the bacterial diversity of atherosclerotic plaques and identified Chryseomonas in all atherosclerotic plaque samples, and Veillonella and Streptococcus in the majority (21). More recently, explorations of the microbiome in the circulating compartment showed that an increase in the proportion of Proteobacteria phylum would predict long-term cardiovascular prognosis (22). The putative role of the blood microbiome in the pathogenesis of other diseases, particularly inflammatory diseases, is beyond the scope of this review and it has been thoroughly reviewed before (23).
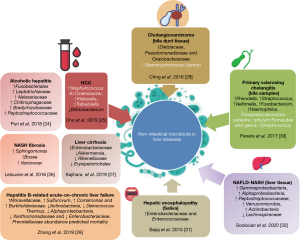
Notably, emerging studies in the field of liver diseases demonstrated the importance of the non-intestinal microbiotas in the development and/or the prognosis of different disease states, from metabolic, inflammatory, and autoimmune diseases to cancer.
These studies involved the profiling of the microbiome of a wide range of non-intestinal biological sources, from samples of the peripheral blood to bile acid and bile duct tissue, saliva, and, more recently, the liver tissue (Figure 2). These investigations lead to important discoveries that generated new hypotheses on host-bacterial interactions. For instance, Puri and coworkers investigated the circulating microbiome signature in patients with alcoholic hepatitis and found that the relative abundance of the phylum Bacteroidetes was significantly decreased in alcohol-consuming patients (24). In addition, the circulating microbiome of subjects consuming alcohol was significantly enriched with the gram-negative bacilli phylum Fusobacteria and its subclasses Fusobacteriales and Leptotrichiaceae (24).
Cho et al. explored the blood microbiome of patients with cirrhosis and hepatocellular carcinoma (HCC) and found that Staphylococcus, Acinetobacter, Klebsiella, and Trabulsiella were significantly enriched in HCC-associated microbiomes (25).
Lelouvier et al. investigated the relationship between the blood microbiota and liver fibrosis in European cohorts of patients with severe obesity and observed that changes in several taxa correlated with the fibrosis status, including the genera Sphingomonas, Bosea, and Variovorax (26).
Kajihara and coworkers examined the peripheral blood microbiota of patients with liver cirrhosis by 16S rRNA gene sequencing and found that while the abundance of Enterobacteriaceae was significantly increased, the abundance of Akkermansia, Rikenellaceae and Erysipelotrichales was decreased in cirrhotic patients (27).
Zhang et al. evaluated the blood microbiome in patients with hepatitis B-related acute-on-chronic liver failure and found that that the relative abundance of Prevotellaceae independently predicted 28-day mortality (28).
On the other hand, there were interesting studies focused on the examination of the microbiome profile of bile acid and/or bile duct tissue samples. For example, Chng and coworkers found that several bacterial families with a significant increase in Stenotrophomonas species allowed for distinguishing tumors vs. paired non-tumors samples in patients with cholangiocarcinoma (29). Pereira et al. collected bile samples from the common bile duct in patients at different stages of primary sclerosing cholangitis and found that Streptococcus abundance was positively correlated with an increase in the disease severity (30).
Based on the profiling of salivary microbiota, Bajaj and coworkers (31) reported that salivary dysbiosis was greater in patients with cirrhosis who developed 90-day hospitalizations because of hepatic encephalopathy.
Finally, we have explored the liver tissue bacterial DNA profile in patients with NAFLD and different phenotypic and histological characteristics and found for the first time that the liver tissue of NAFLD patients contains a diverse repertoire of bacterial DNA (up to 2.5×104 read counts) (32). More importantly, we observed that the liver metataxonomic signature may explain differences in the NAFLD pathogenic mechanisms as well as physiological functions of the host (32).
Biogeography of the liver tissue bacterial DNA of patients with NAFLD
The liver acts as a key filtration system of the whole body by receiving blood from the hepatic artery and the portal vein. Therefore, not only microbes would become entrapped in the complex liver anatomy but, more importantly, bacterial-derived products that are likely to be potentially powerful stimuli for initiating the inflammatory response.
Translocation of bacterial products from the gastrointestinal tract to the liver is largely acknowledged as implicated in the complications of liver cirrhosis (33). More recently, the existence of crosstalk between microbes and host liver cells that control metabolism has been proposed to explain not only de novo hepatic lipogenesis that occurs in NAFLD but also associated liver damage as elegantly reviewed by Tilg et al. (34).
We have profiled the liver tissue microbial DNA in patients with NAFLD and different histological stages (32). Regarding the biogeography of the bacterial DNA found in the liver tissue specimens, we found that the hepatic microbiome profile reflects bacterial DNA communities that can be found not only across the entire gastrointestinal tract—from the oral cavity to the distal colon—but also bacterial DNA potentially derived of other body sites (Figure 3). For example, while we found that many bacterial DNA in the liver tissue samples of patients with NAFLD have been previously reported as microorganisms that inhabit the human intestinal tract, for example, Bifidobacterium, Peptostreptococcus, Stenotrophomonas, Xanthomonadaceae, Lachnoclostridium, and Ruminococcaceae—among many other taxa (35), we observed bacterial DNA potentially derived from other body sites as well. For example, we found that Deinococcus Thermus, which is recognized as one of the most extremophilic phyla of Bacteria (36), was distinctively overabundant in the liver of non-morbidly obese patients with NAFLD (32). Deinococcus Thermus species represent a fraction of the bacterial communities residing in the human stomach (37), which is known for its acidic and inhospitable environment. These results might suggest the potential source of liver bacterial DNA that could migrate from the stomach to the liver. Also, this result might suggest the possibility of different stress-related tissue environments in different cohorts of patients, for example oxidative and/or metabolic stress (38-40), which would, in turn modulate bacterial communities’ adaptation and survival.
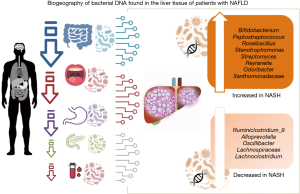
In addition, the findings of our study suggested that some bacterial DNA might be derived from the oral microbiome (41), for example, Actinobacteria and Prevotella, and some species of Deltaproteobacteria.
Other features deserve particular attention. For example, we found the presence of bacterial DNA of Gemella genus in patients with higher scores of portal inflammation (32). Gemella is abundant in the proximal small intestine, and some species of this genus, for instance, Gemella spp can translocate to other organs and thus cause systemic diseases (42).
Likewise, we found that the abundance of bacterial DNA of Peptostreptococcus was significantly increased in the liver of morbidly obese patients with NASH (32). The presence in the gut of members of the Peptostreptococcaceae family is often associated with compromised health, for instance, elderly hospitalized patients with Clostridium difficile infection exposed to previous antibiotic (43), ulcerative colitis (44), and colorectal cancer (45).
Microbiota or bacterial DNA can have negative and positive effects on the host. For instance, we found that depletion of DNA from Lachnospiraceae family members was particularly associated with severe histological features (32). Several members of the Lachnospiraceae family are butyrate producers; microbial butyrate-producing communities are often reported as associated with the healthy microbiota.
Besides, microbiota-derived butyrate presumably presents several health benefits. For example, butyrate is able to beneficially affect oxidative stress in the healthy human colon (46), it might have anti-proliferative effects by interfering with prostaglandin E2 synthesis (47), and butyrate may modulate immunological homeostasis in the gut (48). The decrease in butyrate from gut bacteria has been also linked to energy imbalance and disturbances in body weight and glucose control (49).
A recent experimental study showed that Lachnospiraceae administration attenuates obesity, inflammation, and dysbiosis (50). It has been also reported diminution of Lachnospiraceae microbial populations in patients with inflammatory bowel diseases (51). Remarkably, a human study showed that gut microbial function, inflammation, and cognition were markedly improved in cirrhotic patients by capsular fecal transplant containing Lachnospiraceae (52).
Furthermore, we found that overabundance of DNA derived from gram-negative Proteobacteria, particularly Gammaproteobacteria, was a distinguishing feature of NASH and NASH-related severe histological outcomes (32). For example, DNA derived from Stenotrophomonas was 3.1-fold increased in the liver of morbidly obese patients with NASH. Likewise, DNA derived from Xanthomonadaceae family members was consistently increased in all NASH patients regardless of their body mass index status (32). We also found that DNA from Alpha and Gamma Proteobacteria was consistently associated with increased lobular and portal inflammation scores (32).
Concurrently, we demonstrated not only that the liver tissue of NASH patients contains DNA from Gammaproteobacteria but we found that lipopolysaccharide (LPS)—the major component of the outer membrane in gram-negative bacterial pathogens, predominantly localized in the portal tracts (32).
Bacterial DNA induces inflammatory response
Bacterial DNA is not immunologically inert. Quite the opposite, bacterial DNA is sensed by the immune system as a signal of infectious danger mostly because the structure of bacterial DNA presents high frequency of the dinucleotides CG and lacks of methylation (53).
Interactions between bacterial derived products, including bacterial DNA and RNA induce host inflammatory responses by activating the innate immune defense system, including the inflammasome (54). Activation of innate immune system in response of highly conserved microbial structures is known as pathogen-associated molecular patterns (PAMPs) (55). Collectively, these findings suggest that bacterial DNA itself may be a significant microbial mediator of inflammation in affected tissues by stimulating pro-inflammatory cytokine production, including interferon factor alpha (IFNα) (53) and tumor necrosis factor alpha (TNFα) (56), priming of the inflammasome (57,58), and activation of toll like receptors (59).
Advances in genomics and bioinformatics revealed that bacterial DNA might potentially integrate into the human genome through an RNA intermediate and such integrations are detected more frequently in tumor than normal cells (60). This finding not only transforms current knowledge of host-bacterial-interactions but also opens new avenues of research in the field of NASH and the development of HCC, particularly in non-cirrhotic patients.
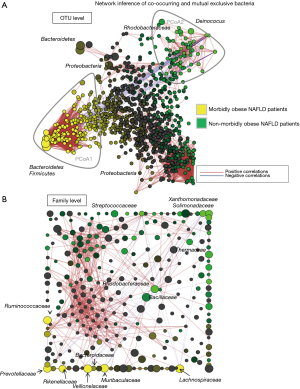
Bacteria-bacteria interactions are another interesting mechanism to explain pathogen-host effects. We hypothesized that differences in liver bacterial DNA diversity between cohorts of NAFLD patients with different phenotypic features, including severely obese versus moderate obese patients, would also reflect putative differences in systemic ecological diversity. We thus performed network analysis of bacterial DNA communities, and observed that bacterial DNA found in the liver of morbidly obese patients with NAFLD clustered separately from the bacterial DNA of the liver of NAFLD-non-morbidly obese patients. More importantly, while clusters of bacterial DNA communities presented significant intra-cluster positive correlations, significant negative co-occurrences between clusters were observed, for example between Gammaproteobacteria and Lachnospiraceae, which were precisely linked to severe and benign histological phenotypes, respectively (Figure 4); this pattern was replicated at all taxonomic levels.
Liver tissue metagenomic findings can be leveraged to identify therapeutic targets against NASH
The putative interplay between the liver tissue cellular components and bacteria-derived products has never been explored before in humans. A rodent study that investigated ethanol-associated compositional microbiome changes along the intestinal tract and the liver showed that ethanol increases bacterial richness in the jejunum and changed bacterial communities in the ileum by inducing a shift towards gram-negative bacteria (61).
As mentioned in the preceding paragraphs, we have shown that the presence of bacterial DNA in the liver tissue of patients with NAFLD would be implicated as a contributing factor in the disease pathogenesis (32).
Microbiome and host genetics are often explored as independent contributing factors in complex disorders, albeit there is a clear and biologically plausible interplay between these two factors. It becomes then obvious that understanding the putative synergistic effect/s between the microbiome and/or its byproducts and the genetic background of the host may serve to predict and design novel personalized treatments. The goal would be to identify novel pathways and interactions with microbial components that could provide effective therapies for NASH. Nevertheless, there are gaps in our understanding of how and which of these putative interactions between the tissue microbiota and the host-genetics impact on the disease biology.
Recently, a study by Abul-Husn et al. revealed that rs72613567, a loss-of-function variation in HSD17B13 (17-beta-hydroxysteroid dehydrogenase 13) gene, confers protection against chronic liver injury and mitigates the progression of NAFLD and alcoholic liver disease in European Americans (62). We further replicated these findings and demonstrated that the rs72613567 A-INS allele protects against NAFLD [odds ratio (OR) per adenine allele =0.667; 95% CI, 0.486–0.916; P=0.012] (63). In addition, we found that the A-INS allele protects against NASH (OR =0.612; 95% CI, 0.388–0.964), ballooning degeneration (OR =0.474; 95% CI, 0.267–0.842), lobular inflammation (OR =0.475; 95% CI, 0.275–0.821), and fibrosis (OR =0.590; 95% CI, 0.361–0.965) (63). Subsequent studies from other groups (64-67) found similar results. Importantly, as shown by immunohistochemistry, there was a dose-dependent decreasing effect of the rs72613567 A-INS on the expression levels of the HSD17B13 protein in the liver (63).
Notably, we have performed the liver transcriptome analysis of NAFLD patients carrying the rs72613567 (–/–) homozygous genotype versus the reference group [rs72613567 (A/A) homozygous genotype], and we found that the most (≥3.5-fold) downregulated and upregulated genes in the liver of patients carrying the rs72613567 (–/–) genotype were in biological pathways related to the immune system, including antigen presentation and interferon-related processes, as well as cytokine signaling (63). Accordingly, previous evidence from experimental studies showed that hydroxysteroid (17 beta) dehydrogenase 13 deficiency triggers hepatic steatosis and inflammation in mice (68). Together, these observations raised the possibility that therapeutic inhibition of HSD17B13 could represent a new strategy to treat NASH patients with more severe liver histology (1,63,69).
HSD17B13 as a hydroxysteroid dehydrogenase (HSD) belongs to the superfamily of short-chain dehydrogenases/reductases (SDR); these enzymes are involved in the steroid hormone metabolism among others. Disruption in the signaling of members of the SDR family may cause a wide range of inflammatory diseases, from hypertension to cancer (70). Likewise, the metabolic activity of microorganisms co-inhabiting the human body, particularly the gut, may act as a parallel endocrine site by producing hormones that can interfere with human physiology. For example, it has been long speculated that the composition of the gastrointestinal microbiota may be involved in the development of cancer (71). Also, interference with steroid hormone metabolism, e.g., progesterone, estradiol and cortisol, may affect the inflammatory response (72).
Hence, it was speculated about a possible relationship between host-genetics and microbiota that would modulate the inflammatory response through to the activity of steroid metabolism, and this relationship might alternatively affect bile acid signaling as well (70).
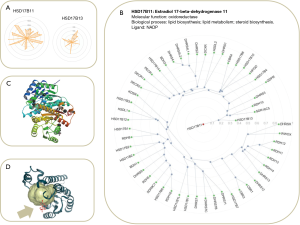
Meanwhile, it remains to be explored whether the rs72613567 variant in HSD17B13 presents any direct or indirect interaction effect with the microbial composition of the body, including the liver or bile acid bacterial composition, which might explain in part the missing heritability of NAFLD and NASH (1). Unfortunately, knowledge of the function and regulation of HSD17B13 is still missing; no crystal structure was deposited in the protein database (PDB) for HSD17B13 either. Therefore, criteria for exploration of HSD17B13 druggability predictions are currently inconclusive. Conversely, HSD17B11 (estradiol 17-beta-dehydrogenase 11)—a protein of the same family that HSD17B13, has been largely studied (Figure 5A). HSD17B13 shares ~65% of its amino acid sequence with HSD17B11 (E value: 1.3e–131) being the closest family member (Figure 5B), and similarly to HSD17B13, HSD17B11 is located in the endoplasmic reticulum. As a proof-of-concept, we modeled the putative druggability (the ability of a binding site to bind a drug-like ligand) of HSD17B11 to demonstrate the potentiality of targeting HSD17B13. Together, the analysis of available HSD17B11 protein structure was straightforward suggesting that proteins in the same family, including HSD17B13, are predicted to be druggable (Figure 5C,D). Analysis of structural ligandability of HSD17B11 (Figure 5C,D) shows a binding site conformation with a putative pocket for binding small drugs.
Variants in many other candidate genes remain unexplored, for example rs59846192 in DMRTB1 (DMRT like family B with proline rich C-terminal 1) that has been associated with relative abundance of Lachnospira in earlier studies (73).
Nevertheless, the most relevant message is that microbial associations with host-genome variants are body site-specific (74). Hence, associations of host-genome variants with the fecal microbiome do not necessarily explain the effect of liver tissue microbial and host-genetics interactions.
Areas of future research
An important yet poorly explored aspect of the microbial and host-genetics interactions is the possibility of drug-metabolizing proteins encoded by the microbiome genes might interfere with patients’ response to medicinal drugs. There is evidence from recent literature that intestinal metagenomic abundance of bacterial phyla, genera, species, and their gene products are able to interfere with many drugs used to treat human diseases, for instance norethindrone acetate and famciclovir (75). A study that combined genomics of human gut commensals and gnotobiotic animal experiments to quantify microbiota and host contributions to drug metabolism provided remarkable evidence on the microbiome drug-metabolizing activity (76). The model consisted of eight compartments, including three sections of the small intestine, three sections of the large intestine, gallbladder, and a central compartment, representing processes occurring in serum, liver and other tissues (76). This investigation demonstrated that while bacterial drug conversion influences both intestinal and systemic drug metabolite levels, the main factors affecting systemic drug metabolite levels were systemic metabolite elimination and biliary secretion rates of the drug (76). Surprisingly, drug metabolites produced by the gut microbiota substantially contributed to systemic exposure (76). It remains to be explored whether tissular metataxonomic profiles represent microbiotas. Tissular microbiotas, in particular the liver, might affect drug pharmacokinetics, possessing potential implications in novel discoveries for the treatment of NASH. Deciphering the interrelationship between liver tissular microbiota and anti-NASH drugs pharmacokinetics and pharmacodynamics would be a breakthrough in the understanding of interpersonal variability in drug response.
Acknowledgments
Funding: This study was partially supported by grants PID-C2012-0061, PICT 2014-0432, PICT 2014-1816, PICT 2015-0551, and PICT 2016-0135 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT), CONICET Proyectos Unidades Ejecutoras 2017, PUE 0055.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://hbsn.amegroups.org/article/view/10.21037/hbsn-20-270/coif). SS and CJP serve as unpaid editorial board members of Hepatobiliary Surgery and Nutrition.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sookoian S, Pirola CJ. Genetics of nonalcoholic fatty liver disease: from pathogenesis to therapeutics. Semin Liver Dis 2019;39:124-40. [Crossref] [PubMed]
- Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015;1:15080. [Crossref] [PubMed]
- Friedman SL, Neuschwander-Tetri BA, Rinella M, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908-22. [Crossref] [PubMed]
- Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764-75. [Crossref] [PubMed]
- Caussy C, Tripathi A, Humphrey G, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun 2019;10:1406. [Crossref] [PubMed]
- Da Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep 2018;8:1466. [Crossref] [PubMed]
- Duarte SMB, Stefano JT, Miele L, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. Nutr Metab Cardiovasc Dis 2018;28:369-84. [Crossref] [PubMed]
- Hoyles L, Fernandez-Real JM, Federici M, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med 2018;24:1070-80. [Crossref] [PubMed]
- Jiang W, Wu N, Wang X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep 2015;5:8096. [Crossref] [PubMed]
- Kim HN, Joo EJ, Cheong HS, et al. Gut microbiota and risk of persistent nonalcoholic fatty liver diseases. J Clin Med 2019;8:1089. [Crossref] [PubMed]
- Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab 2017;25:1054-62.e5. [Crossref] [PubMed]
- Michail S, Lin M, Frey MR, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol 2015;91:1-9. [Crossref] [PubMed]
- Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013;58:120-7. [Crossref] [PubMed]
- Schwimmer JB, Johnson JS, Angeles JE, et al. Microbiome signatures associated with steatohepatitis and moderate to severe fibrosis in children with nonalcoholic fatty liver disease. Gastroenterology 2019;157:1109-22. [Crossref] [PubMed]
- Shen F, Zheng RD, Sun XQ, et al. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int 2017;16:375-81. [Crossref] [PubMed]
- Wong VW, Tse CH, Lam TT, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One 2013;8:e62885 [Crossref] [PubMed]
- Zhang Y, Xu J, Wang X, et al. Changes of intestinal bacterial microbiota in coronary heart disease complicated with nonalcoholic fatty liver disease. BMC Genomics 2019;20:862. [Crossref] [PubMed]
- Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 2013;57:601-9. [Crossref] [PubMed]
- Ponziani FR, Bhoori S, Castelli C, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019;69:107-20. [Crossref] [PubMed]
- Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694-7. [Crossref] [PubMed]
- Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A 2011;108:4592-8. [Crossref] [PubMed]
- Amar J, Lange C, Payros G, et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PLoS One 2013;8:e54461 [Crossref] [PubMed]
- Potgieter M, Bester J, Kell DB, et al. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev 2015;39:567-91. [Crossref] [PubMed]
- Puri P, Liangpunsakul S, Christensen JE, et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 2018;67:1284-302. [Crossref] [PubMed]
- Cho EJ, Leem S, Kim SA, et al. Circulating microbiota-based metagenomic signature for detection of hepatocellular carcinoma. Sci Rep 2019;9:7536. [Crossref] [PubMed]
- Lelouvier B, Servant F, Paisse S, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology 2016;64:2015-27. [Crossref] [PubMed]
- Kajihara M, Koido S, Kanai T, et al. Characterisation of blood microbiota in patients with liver cirrhosis. Eur J Gastroenterol Hepatol 2019;31:1577-83. [Crossref] [PubMed]
- Zhang Y, Zhao R, Shi D, et al. Characterization of the circulating microbiome in acute-on-chronic liver failure associated with hepatitis B. Liver Int 2019;39:1207-16. [Crossref] [PubMed]
- Chng KR, Chan SH, Ng AHQ, et al. Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine 2016;8:195-202. [Crossref] [PubMed]
- Pereira P, Aho V, Arola J, et al. Bile microbiota in primary sclerosing cholangitis: Impact on disease progression and development of biliary dysplasia. PLoS One 2017;12:e0182924 [Crossref] [PubMed]
- Bajaj JS, Betrapally NS, Hylemon PB, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 2015;62:1260-71. [Crossref] [PubMed]
- Sookoian S, Salatino A, Castaño GO, et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut 2020;69:1483-91. [Crossref] [PubMed]
- Wiest R, Garcia-Tsao G. Bacterial translocation (BT) in cirrhosis. Hepatology 2005;41:422-33. [Crossref] [PubMed]
- Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut 2016;65:2035-44. [Crossref] [PubMed]
- Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014;38:996-1047. [Crossref] [PubMed]
- Theodorakopoulos N, Bachar D, Christen R, et al. Exploration of Deinococcus-Thermus molecular diversity by novel group-specific PCR primers. Microbiologyopen 2013;2:862-72. [PubMed]
- Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103:732-7. [Crossref] [PubMed]
- Sookoian S, Flichman D, Scian R, et al. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol 2016;240:437-49. [Crossref] [PubMed]
- Sookoian S, Castano GO, Scian R, et al. Heat shock protein 27 is down-regulated in ballooned hepatocytes of patients with nonalcoholic steatohepatitis (NASH). Sci Rep 2016;6:22528. [Crossref] [PubMed]
- Sookoian S, Castano GO, Scian R, et al. Serum aminotransferases in nonalcoholic fatty liver disease are a signature of liver metabolic perturbations at the amino acid and Krebs cycle level. Am J Clin Nutr 2016;103:422-34. [Crossref] [PubMed]
- Dewhirst FE. The oral microbiome: critical for understanding oral health and disease. J Calif Dent Assoc 2016;44:409-10. [PubMed]
- Ou G, Hedberg M, Horstedt P, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol 2009;104:3058-67. [Crossref] [PubMed]
- Milani C, Ticinesi A, Gerritsen J, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep 2016;6:25945. [Crossref] [PubMed]
- Rajilić-Stojanović M, Shanahan F, Guarner F, et al. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 2013;19:481-8. [Crossref] [PubMed]
- Chen W, Liu F, Ling Z, et al. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 2012;7:e39743 [Crossref] [PubMed]
- Hamer HM, Jonkers DM, Bast A, et al. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr 2009;28:88-93. [Crossref] [PubMed]
- Gostner A, Dusel G. Kelber Eet al. Comparisons of the anti-proliferative effects of butyrate and aspirin on human colonic mucosa in vitro. Eur J Cancer Prev 2000;9:205-11. [Crossref] [PubMed]
- Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446-50. [Crossref] [PubMed]
- De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014;156:84-96. [Crossref] [PubMed]
- Truax AD, Chen L, Tam JW, et al. The inhibitory innate immune sensor NLRP12 maintains a threshold against obesity by regulating gut microbiota homeostasis. Cell Host Microbe 2018;24:364-78.e6. [Crossref] [PubMed]
- Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780-5. [Crossref] [PubMed]
- Bajaj JS, Salzman N, Acharya C, et al. Microbial functional change is linked with clinical outcomes after capsular fecal transplant in cirrhosis. JCI Insight 2019;4:e133410 [Crossref] [PubMed]
- Heeg K, Sparwasser T, Lipford GB, et al. Bacterial DNA as an evolutionary conserved ligand signalling danger of infection to immune cells. Eur J Clin Microbiol Infect Dis 1998;17:464-9. [Crossref] [PubMed]
- Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol 2009;19:455-64. [Crossref] [PubMed]
- Eigenbrod T, Dalpke AH. Bacterial RNA: an underestimated stimulus for innate immune responses. J Immunol 2015;195:411-8. [Crossref] [PubMed]
- Sweet MJ, Stacey KJ, Kakuda DK, et al. IFN-gamma primes macrophage responses to bacterial DNA. J Interferon Cytokine Res 1998;18:263-71. [Crossref] [PubMed]
- Franchi L, Munoz-Planillo R, Reimer T, et al. Inflammasomes as microbial sensors. Eur J Immunol 2010;40:611-5. [Crossref] [PubMed]
- Sun L, Wu J, Du F, Chen X, et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013;339:786-91. [Crossref] [PubMed]
- Hemmi H, Takeuchi O, Kawai T, et al. A toll-like receptor recognizes bacterial DNA. Nature 2000;408:740-5. [Crossref] [PubMed]
- Riley DR, Sieber KB, Robinson KM, et al. Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput Biol 2013;9:e1003107 [Crossref] [PubMed]
- Bluemel S, Wang L, Kuelbs C, et al. Intestinal and hepatic microbiota changes associated with chronic ethanol administration in mice. Gut Microbes 2020;11:265-75. [Crossref] [PubMed]
- Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med 2018;378:1096-106. [Crossref] [PubMed]
- Pirola CJ, Garaycoechea M, Flichman D, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res 2019;60:176-85. [Crossref] [PubMed]
- Di Sessa A, Umano GR, Cirillo G, et al. The rs72613567: TA variant in the hydroxysteroid 17-beta dehydrogenase 13 gene reduces liver damage in obese children. J Pediatr Gastroenterol Nutr 2020;70:371-4. [Crossref] [PubMed]
- Ma Y, Belyaeva OV, Brown PM, et al. 17-beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology 2019;69:1504-19. [Crossref] [PubMed]
- Stickel F, Lutz P, Buch S, et al. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology 2020;72:88-102. [Crossref] [PubMed]
- Yang J, Trepo E, Nahon P, et al. A 17-beta-hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology 2019;70:231-40. [Crossref] [PubMed]
- Adam M, Heikela H, Sobolewski C, et al. Hydroxysteroid (17beta) dehydrogenase 13 deficiency triggers hepatic steatosis and inflammation in mice. FASEB J 2018;32:3434-47. [Crossref] [PubMed]
- Sookoian S, Arrese M, Pirola CJ. Genetics meets therapy? Exome-wide association study reveals a loss-of-function variant in 17-beta-hydroxysteroid dehydrogenase 13 that protects patients from liver damage and nonalcoholic fatty liver disease progression. Hepatology 2019;69:907-10. [Crossref] [PubMed]
- Kisiela M, Skarka A, Ebert B, et al. Hydroxysteroid dehydrogenases (HSDs) in bacteria: a bioinformatic perspective. J Steroid Biochem Mol Biol 2012;129:31-46. [Crossref] [PubMed]
- Drasar BS, Hill MJ. Intestinal bacteria and cancer. Am J Clin Nutr 1972;25:1399-404. [Crossref] [PubMed]
- Clark DT, Soory M. The metabolism of cholesterol and certain hormonal steroids by Treponema denticola. Steroids 2006;71:352-63. [Crossref] [PubMed]
- Turpin W, Espin-Garcia O, Xu W, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 2016;48:1413-7. [Crossref] [PubMed]
- Kolde R, Franzosa EA, Rahnavard G, et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med 2018;10:6. [Crossref] [PubMed]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, et al. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019;570:462-7. [Crossref] [PubMed]
- Zimmermann-Kogadeeva M, Zimmermann M, Goodman AL. Insights from pharmacokinetic models of host-microbiome drug metabolism. Gut Microbes 2020;11:587-96. [Crossref] [PubMed]