
Nonalcoholic steatohepatitis pharmacotherapy and predictors of response: dual role of aminotransferases as biosensors of metabolism and biomarkers of histological improvement
Nonalcoholic steatohepatitis (NASH)—the severe histological form of nonalcoholic fatty liver disease (NAFLD)—is regarded as a major health problem worldwide (1,2). The disease can progress to liver cirrhosis and eventually to hepatocellular carcinoma (1,3). Treatment of NASH and NASH-fibrosis is thus increasingly being given priority in the clinical field. Yet, these efforts face challenges not only related to the design and validation of novel pharmacological agents, but also to the need to optimize treatment options as well as improve access to therapies. In fact, while many drugs are currently being tested for safety and efficacy, only a few approved options for the treatment of NASH presently exist (4,5). It is thus expected that, in the near future, emphasis will be given to selecting the right drug for treating NASH patients from a wide spectrum of treatment choices. Hence, identification of early predictors of treatment response represents an unmet and relevant need.
Loomba and coworkers have recently published results yielded by a post-hoc analysis of clinical predictors of histologic response (6), whereby the data was sourced from the FLINT trial—a double-blind, placebo-controlled, randomized clinical trial conducted at multiple medical centers across the U.S. involving non-cirrhotic NASH patients—to assess the treatment efficacy of obeticholic acid (OCA, a farnesoid X receptor agonist) given orally (25 mg daily) for 72 weeks (7). Specifically, the authors aimed at identifying baseline and early on-treatment factors that might predict histologic response. By utilizing a multivariable-adjusted model, the authors found that baseline NAS (NASH activity score) >5, baseline triglyceride levels ≤154 mg/dL, baseline international normalized ratio ≤1, baseline aspartate aminotransferase (AST) level ≤49 IU/L, and a decrease in alanine aminotransferase (ALT) levels at week 24 by at least 17 IU/L were significantly associated with histological improvement (6). These interesting and promising findings deserve some reflections.
What is the biological meaning of these findings?
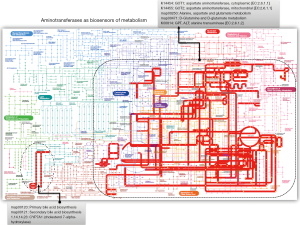
Notably, all predictors in the model with the exception of the NAS score are indeed biochemical measurements that include liver enzymes, specifically aminotransferases, and circulating levels of triglycerides. One might argue that liver enzymes are expected predictors of treatment response because ALT and AST have been widely used in prior studies not only as surrogate biomarkers of liver disease severity (8,9) but also to monitor response to any liver disease, particularly the treatment of chronic viral hepatitis. Nevertheless, more recent data indicate that serum concentrations aminotransferases are linked to cardiovascular disease and the metabolic syndrome (9-11). Therefore, it has been postulated that elevated aminotransferase values approaching the upper-normal threshold reflect high levels of hepatic transamination in response to increased NAFLD-associated metabolic demands (9,11). In fact, we have evaluated the expression levels of aminotransferases in the liver of patients with NAFLD and have explored their relationship with global metabolic changes in the circulation (11). Profiling of circulating metabolites indicated that serum aminotransferase concentrations are a signature of liver metabolic perturbations, particularly at the amino acid metabolism and Krebs cycle level (11). Robust confirmation of that finding can be found in the representation of GPT and GOT—including their isoenzymes—in the global metabolic map (Figure 1). The pathways in which aminotransferases are involved cover not only an extensive portion of the carbohydrate and amino acid metabolism, but energy metabolism as well (Figure 1). The role of GOT1 [aspartate aminotransferase, cytoplasmic (EC:2.6.1.1)] is particularly interesting, as it presents 8 direct hits and 256 indirect hits in the global metabolic map (Figure 1). Consequently, it is plausible to suggest that baseline levels of AST—as reported by Loomba and coworkers (6)—are not necessarily prognostic indicators of histological improvement but also dynamic biosensors of the patients’ metabolic status.
Clinical predictors versus genetic predictors of treatment response
Another important issue to discuss is the potentiality for including other factors, such as genetic predictors, in the model. This hypothesis could be tested, for example, via a genome-wide exploration of variants (GWAS), by necessity in a large cohort. Another, more feasible, alternative could be a rational selection of gene candidates based on biological modeling. We explored the connectivity network around NR1H4 (FXR) and aminotransferase genes (GPT1, GPT2, GOT1, and GOT2) and found some potential candidates for inclusion into the model, including CYP7A1 (cytochrome P450 family 7 subfamily A member 1) (Figure 2A). This finding is not unexpected, as FXR not only regulates bile acid biosynthesis controlling CYP7A1 transcription through the FGFR4-JNK-HNF4α pathway, but also hepatic gluconeogenesis, glycolisis and lipogenesis, including triglyceride biosynthesis through the SHP (the inhibitory nuclear receptor small heterodimer partner)-SREBP-1c pathway (12). This complex molecular interaction of the drug target may explain some adverse effects of OCA on insulin resistance (7), prompting the question of how the incorporation of plasma glucose-related variables may affect the predictive model results reported by Loomba et al. (6). As a proof of principle, it can be shown that variants in, or in close proximity to, CYP7A1 are associated with lipid traits in the UK Biobank (Figure 2B), which reinforces the concept that baseline levels of lipids in patients with NASH that responded or not to OCA are genetically modulated. In addition, text mining would suggest that GPT (ALT) could be a central node in connecting the sub-network of nuclear receptors with the one composed by aminotransferases and enzymes of the glutamic acid metabolism, as GPT and FXR might be co-expressed in some species (Figure 2B).
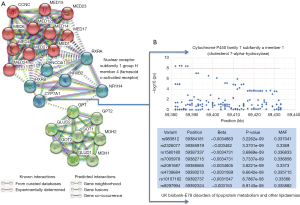
A final question that still remains unanswered pertains to the generalizability of Loomba et al.’s work (6). Specifically, one may wonder whether the reported findings can be extended to all populations and to all kinds of anti-NASH drugs. Finally, as FXR and CYP7A1 are at the core of the metabolism of bile acids, it remains to be explored whether other environmental factors, such as diet or host “microbiotas”, may modulate the therapeutic response of any drug but OCA in particular, and vice versa (13).
Acknowledgments
Funding: Agencia Nacional de Promoción Científica y Tecnológica, Fondo para la Investigación Científica y Tecnológica (FonCyT) (PICT 2014-0432 and PICT 2015-0551 to S Sookoian and PICT 2014-1816 and 2016-0135 to CJ Pirola).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015;1:15080. [Crossref] [PubMed]
- Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328-57. [Crossref] [PubMed]
- Friedman SL, Neuschwander-Tetri BA, Rinella M, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908-22. [Crossref] [PubMed]
- Konerman MA, Jones JC, Harrison SA. Pharmacotherapy for NASH: Current and emerging. J Hepatol 2018;68:362-75. [Crossref] [PubMed]
- Pirola CJ, Sookoian S. Tackling the complexity of nonalcoholic steatohepatitis treatment: challenges and opportunities based on systems biology and machine learning approaches. Hepatobiliary Surg Nutr 2018;7:495-8. [Crossref] [PubMed]
- Loomba R, Sanyal AJ, Kowdley KV, et al. Factors Associated With Histologic Response in Adult Patients With Nonalcoholic Steatohepatitis. Gastroenterology 2019;156:88-95.e5. [Crossref] [PubMed]
- Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet 2015;385:956-65. [Crossref] [PubMed]
- Sherman KE. Alanine aminotransferase in clinical practice. A review. Arch Intern Med 1991;151:260-5. [Crossref] [PubMed]
- Sookoian S, Pirola CJ. Alanine and aspartate aminotransferase and glutamine-cycling pathway: their roles in pathogenesis of metabolic syndrome. World J Gastroenterol 2012;18:3775-81. [Crossref] [PubMed]
- Porter SA, Pedley A, Massaro JM, et al. Aminotransferase levels are associated with cardiometabolic risk above and beyond visceral fat and insulin resistance: the Framingham Heart Study. Arterioscler Thromb Vasc Biol 2013;33:139-46. [Crossref] [PubMed]
- Sookoian S, Castaño GO, Scian R, et al. Serum aminotransferases in nonalcoholic fatty liver disease are a signature of liver metabolic perturbations at the amino acid and Krebs cycle level. Am J Clin Nutr 2016;103:422-34. [Crossref] [PubMed]
- Staels B, Handelsman Y, Fonseca V. Bile acid sequestrants for lipid and glucose control. Curr Diab Rep 2010;10:70-7. [Crossref] [PubMed]
- Friedman ES, Li Y, Shen TD, et al. FXR-Dependent Modulation of the Human Small Intestinal Microbiome by the Bile Acid Derivative Obeticholic Acid. Gastroenterology 2018;155:1741-52.e5. [Crossref] [PubMed]


