DGAT1-deficiency affects the cellular distribution of hepatic retinoid and attenuates the progression of CCl4-induced liver fibrosis
Introduction
Retinoids (vitamin A and its natural metabolites and synthetic analogs) are potent transcriptional regulators (1-3) affecting expression levels of more than 500 genes (3). The all-trans- and 9-cis-isomers of retinoic acid are transcriptionally active retinoid metabolites, regulating transcription upon binding to one of 3 distinct retinoic acid receptors (RARs) or 3 distinct retinoid X receptors (RXRs) (1-3). There is growing understanding that retinoids also act within cells to regulate signal transduction pathways independent of their interactions with RARs and RXRs (4,5). Retinoids participate in regulating normal cellular processes including differentiation, proliferation, and apoptosis (6,7) and are therefore needed for maintaining immunity, male and female reproduction, pre- and post-natal development, barrier function, and vision (6,7). There is also considerable interest in the study of retinoid actions in both the prevention and the development of metabolic disease including obesity, insulin resistance, liver disease and cardiovascular disease (8-14).
Although the isomers of retinoic acid represent the functionally important retinoid species, the most abundant retinoids in the body are retinyl esters, followed quantitatively by retinol (15). Collectively, retinyl esters and retinol account for greater than 95% of all the retinoid present in the body. It has been estimated that 80-90% of the retinoid in the body of healthy, well-nourished mammals is stored in the liver. The majority of this retinoid is stored as retinyl ester within the lipid droplets of hepatic stellate cells (HSCs), with the remainder of hepatic retinyl ester and retinol found in hepatocytes (16,17). It is well established that, upon hepatic injury, HSC retinyl ester stores are rapidly lost as the HSCs become activated and assume the characteristics of fibrogenic, proliferative myofibroblasts (17). At present, it remains unclear if this concomitant loss of retinyl ester during HSC activation contributes to the progression of fibrogenesis, although the absence of HSC retinyl ester stores in mice lacking the enzyme lecithin:retinol acyltransferase (LRAT) does not affect the development of CCl4-induced hepatic fibrosis (18).
LRAT catalyzes the transesterification of the sn-1 acyl moiety of membrane phosphatidyl choline to retinol in the synthesis of retinyl ester (19). Studies of induced mutant mice have established that LRAT is responsible for almost all retinyl ester formation within the body (20-22). The older literature indicates that an acyl CoA-dependent enzyme termed acyl CoA:retinol acyltransferase (ARAT) also contributes to retinyl ester formation in liver and intestine (17,23). Diacylglycerol O-acyltransferase 1 (DGAT1) is one of two mammalian enzymes, the other being diacylglycerol O-acyltransferase 2 (DGAT2), that catalyze acyl CoA-dependent triglyceride formation from diacylglycerol (24). DGAT1, but not DGAT2, possesses ARAT activity in vitro (21,25,26) and can act in vivo as an ARAT in small intestine and skin (27,28). Although DGAT1 is expressed in both HSCs and hepatocytes, based on data obtained from Lrat-deficient mice, DGAT1 does not catalyze retinyl ester formation in either HSCs or hepatocytes, since these mice completely lack hepatic retinyl esters and HSC lipid droplets (21). This is true even when the animals were fed very high levels of dietary retinoid (29). These findings are surprising since DGAT1 catalyzes retinyl ester formation in some tissues and, based on the studies of whole liver homogenates, may be able to catalyze acyl CoA-dependent retinyl ester synthesis in liver (21,25-28). It has been reported by Yamaguchi et al. (30) that the inhibition of DGAT1 in primary rat HSCs with anti-sense oligonucleotides reduced mRNA expression levels of markers for HSC activation. These investigators further reported elevated cellular retinol-binding protein, type 1 (Rbp1) and Lrat mRNA expression in anti-sense treated HSCs and proposed that the loss of DGAT1, as an important ARAT activity in the liver, leaves more retinol available for oxidation to retinoic acid (30). The present studies explore in greater depth the role of DGAT1 in hepatic retinoid metabolism and HSC activation.
Materials and methods
Animals, animal husbandry and diets
Mice employed in this study were on the C57BL/6J genetic background. Diet-matched male Dgat1–/– and WT mice were 6 months of age at the time of experiments. Throughout life, mice had ad libitum access to water and a standard, nutritionally complete rodent chow diet containing 15 IU retinol/g diet (W. F. Fisher and Sons, Inc., Somerville, USA). The mice were maintained on a 12-h dark-light cycle in a conventional barrier facility with the period of darkness between 7:00 p.m. and 7:00 a.m. All experiments were conducted in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals [National Research Council 2011. Guide for the Care and Use of Laboratory Animals (8th ed), Natl Acad Press, Washington, USA], and were approved by the Columbia University Institutional Animal Care and Use Committee.
Isolation of hepatocytes
For primary hepatocyte isolations, livers were perfused in situ with 1 mM EDTA for 5 min followed by 0.5 mg/mL collagenase D (Roche Diagnostics, Indianapolis, USA) for 13 min. Liver digests were filtered through a 100 µm nylon cell strainer (BD Biosciences) to remove undigested material and centrifuged at 20 ×g for 5 min to sediment hepatocytes. Non-parenchymal cells and debris were removed with the supernatant and the hepatocytes subsequently were washed 4 times with Gey’s Balanced Salt Solution (GBSS, Sigma, St. Louis, MO), centrifuging at 20 ×g for 5 min, 50 ×g for 10 min, and 2 times at 20 ×g for 5 min. Purified hepatocytes were flash frozen in liquid N2 and stored as pellets at –80 °C until further analysis.
Isolation and culture of HSCs
Primary HSCs were isolated from WT and Dgat1–/– mice according to established protocols (31-33). Livers were perfused sequentially in situ with 0.5 mM EGTA for 5 min, pronase E (0.4 mg/mL, EMD Chemicals Inc., Gibbstown, USA) for 5 min, and collagenase D (0.5 mg/mL, Roche Diagnostics) for 8 min, all at a flow rate of 5 mL/min. The resulting liver digest was filtered through a 100 µm nylon cell strainer (BD Biosciences) and washed twice with GBSS containing DNase I (2 mg/mL, Roche Diagnostics) and by centrifugation at 580 ×g for 10 min. HSCs were purified by flotation through 9% (w/v) Nycodenz (Axis-Shield PoC AS, Oslo, Norway) in GBSS lacking NaCl by centrifugation at 1,380 ×g for 15 min. In order to obtain pure samples, all of the primary HSC isolates were subjected to a second flotation step in 9% Nycodenz. The HSCs were then washed with GBSS and either pelleted, flash frozen in liquid N2, and saved in –80 °C for further analysis or resuspended in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, USA) and placed in culture.
For culture, freshly isolated HSCs were plated onto 35 mm plastic dishes at a seeding density of 2.5×105 cells/well. The cells were maintained in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (final concentration 100 units penicillin and 0.1 mg streptomycin/mL) (Sigma Aldrich, St. Louis, USA). Plates were incubated at 37 °C in a humidified atmosphere containing 5% CO2. After 4 h of incubation, the medium was changed to remove dead cells and debris. The next day, the medium was changed again and every 2 days afterwards. For microscopy, freshly isolated HSCs were seeded in 35 mm glass dishes (MatTek Co., Ashland, USA) and images were captured using a FSX100 microscope (Olympus America Inc., Center Valley, USA).
High performance liquid chromatography (HPLC) analysis of retinoids
Tissue and cell retinol and retinyl ester levels were determined as previously described (21). Retinoid extracted in hexane was resuspended in benzene and separated by reversed-phase HPLC using acetonitrile:methanol:methylene chloride (70:15:15, v/v) as running solvent. Retinoid was detected at 325 nm and quantitated by comparing integrated peak areas against a known amount of a purified standard.
Triglyceride analysis
Triglyceride was extracted with chloroform:methanol (2:1, v/v), dried under nitrogen gas, and resuspended in 1% Triton X-100. Triglyceride concentrations were measured using a colorimetric assay kit from Thermo Scientific according to the manufacturer’s instructions.
DGAT enzymatic assay
Total DGAT activity was measured as described in Liu et al. (34) using diacylglycerol and [14C]palmitoyl-CoA as substrates in a first order reaction with respect to protein and reaction time. The triglyceride synthesized was separated by thin layer chromatography on silica plates (Whatman, Maidstone, UK) using hexane:diethyl ether:acetic acid (80:20:1, v/v/v) as solvent and the corresponding band scraped for radioactivity measurement using a liquid scintillation counter (Beckman Coulter, Jersey City, USA).
Hepatic clearance of an oral retinol challenge
The in vivo uptake of recently ingested oral retinol into hepatic cell fractions was assessed using 1 µCi [3H]retinol as radiolabeled tracer, administered in a single gavage dose containing 6 µg of retinol dissolved in 100 µL of corn oil. Hepatocytes and HSCs were isolated 24 h after gavage, as described above. Immediately prior to perfusion of the liver, a small lobe was ligated and excised for whole liver measurement of 3H-cpm. Total retinoids were extracted from the liver and from the isolated cell fractions into chloroform:methanol (2:1, v/v), dried under nitrogen, and measured for radioactivity using a liquid scintillation counter (Beckman Coulter). 3H-cpm detected for the individual cell fractions were normalized to cell number and expressed as a percent of total liver 3H-cpm. These calculations were based on 135 million hepatocytes (35) and 10 million HSCs per gram of mouse liver (36-38).
CCl4 treatment
Two studies were carried out which differed in the doses and durations of CCl4 treatment. For study 1, mice were given an intraperitoneal (IP) injection, every 48 h, of either 1 µL CCl4 per gram of body weight dissolved in corn oil (1:4, v/v) or an equal volume of pure corn oil as control, for a total of four injections. The mice were then euthanized 24 h following the last injection, and liver samples were collected for histology, analysis of mRNA expression and retinoid measurements. For study 2, mice were given IP injections, every 7 days, of either 0.5 µL CCl4 per gram of body weight dissolved in corn oil (1:4, v/v) or an equal volume of corn coil as control, for a total of four injections. Treated mice were euthanized 7 days after the last injection for liver tissues.
Histological analysis
Liver was fixed overnight in 10% formalin and embedded in paraffin for sectioning and Masson’s Trichrome staining. Histology was performed by the Pathology Core Facility at Columbia University Medical Center, and images were captured with a FSX100 biological microscope (Olympus America Inc., Center Valley, USA).
RNA isolation, reverse transcription, and qualitative real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions and treated with DNAse using RNeasy columns (Qiagen, Valencia, USA). cDNA was synthesized with a high-capacity reverse transcription kit (Applied Biosystems, Carlsbad, USA) and subjected to qRT-PCR using SYBR Green PCR master mix from Roche Diagnostics. mRNA levels were quantitated against a standard curve generated by serial dilution of tissue/cell type appropriate cDNA and normalized to 18S rRNA levels.
Statistical analyses
All data are presented as mean ± standard deviation. Statistical differences were determined by student’s t-test, with a P value ˂0.05 indicating statistical significance.
Results
DGAT1-deficiency results in an altered cellular distribution of hepatic retinoid
Primary HSCs were isolated from 6-month-old, male Dgat1–/– and WT mice and analyzed for retinoid content by reversed-phase HPLC. Cellular levels of retinyl esters were significantly elevated in Dgat1–/– compared to WT HSCs (Figure 1A). This was unexpected since total hepatic retinoid levels are not different for Dgat1–/– and WT mice (27). When we analyzed freshly isolated hepatocytes, we found a correspondingly lower retinyl ester content in Dgat1–/– hepatocytes (Figure 1B). Cellular levels of retinol were 2-fold greater in Dgat1–/– compared to WT HSCs (Figure 1C) but were not different between Dgat1–/– and WT hepatocytes (Figure 1D). A similar cellular difference in retinoid distribution was observed 24 h after administration of an oral dose of 6 µg of retinol and 1 µCi [3H]retinol, dissolved in 100 µL of corn oil. HSCs isolated from the Dgat1–/– mice contained significantly more of the radiolabeled dose than WT HSCs (Figure 1E), whereas hepatocytes isolated from Dgat1–/– mice contained less of the dose than WT hepatocytes (Figure 1F). Collectively, these data suggest that, in the absence of DGAT1, retinol is preferentially accumulated by HSCs at the expense of the hepatocyte.
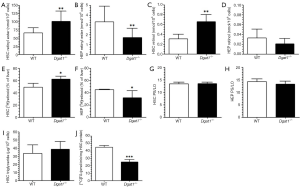
In order to rule out the possibility that the ARAT activity of DGAT1 might contribute to retinol esterification and hence the storage of retinyl ester in hepatocytes, we compared the ratios of the major products of LRAT activity, retinyl palmitate and retinyl stearate, to the major products of ARAT activity, retinyl linoleate and retinyl oleate (27). Identical ratios of these products were observed for Dgat1–/– and WT HSCs (Figure 1G) and hepatocytes (Figure 1H). This rules out DGAT1 as a significant ARAT in these two hepatic cell types. Interestingly, despite the role of DGAT1 in triglyceride synthesis, triglyceride levels were not different in Dgat1–/– HSCs (Figure 1I). This was despite a significant reduction, by approximately 40%, of the total DGAT activity measured in whole Dgat1–/– HSC lysates (Figure 1J). The residual total DGAT activity present in Dgat1–/– HSCs must reflect DGAT2 activity. We observed no differences in Dgat2 mRNA expression between Dgat1–/– and WT HSCs (Table 1).
Full table
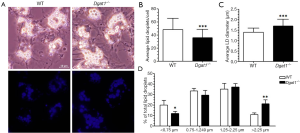
Dgat1–/– HSCs have fewer but larger lipid droplets
Overnight cultures of freshly isolated HSCs from Dgat1–/– and WT mice show visible differences in cell morphology associated with their characteristic lipid droplets. As seen in Figure 2A, the lipid droplets present in Dgat1–/– HSCs appear to be fewer in number but larger than those of WT HSCs. This is also evident in the bottom panels of Figure 2A, which show this same difference in the autofluorescence of endogenous retinoid emitted upon excitation at 330 nm. These morphological differences were analyzed quantitatively by counting the number of lipid droplets present per HSC (Figure 2B) and by measuring the diameter of each of the lipid droplets (Figure 2C). More than 50 cells from each genotype were selected at random from HSC cultures prepared from 4 Dgat1–/– and 5 WT mice. On average, Dgat1–/– HSCs had 36 lipid droplets per cell, while WT HSCs had 49. The WT lipid droplets had an average diameter of 1.4±0.1 µm, while Dgat1–/– lipid droplets were significantly bigger with an average diameter of 1.7±0.1 µm. These values indicate that the total lipid droplet volume is 32% greater in Dgat1–/– compared to WT HSCs, which is quantitatively consistent with the difference in HSC retinyl ester levels observed for the two genotypes (see Figure 1A). When the lipid droplets were grouped according to size, it is clear that there is a shift in the size distribution of the Dgat1–/– lipid droplets (Figure 2D). Compared to WT, in Dgat1–/– HSCs, the larger lipid droplets >2.25 µm in diameter constitute a significantly greater percentage of the total lipid droplets present (21% vs. 11%), whereas the smaller lipid droplets <0.75 µm in diameter make up a significantly smaller percentage (12% vs. 20%).
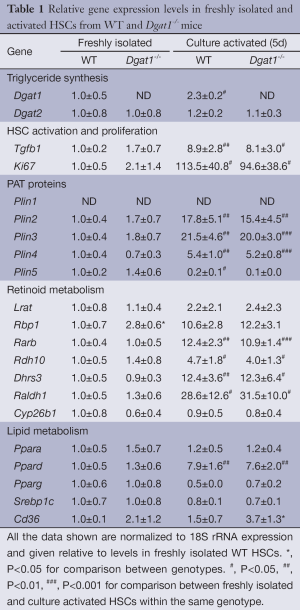
To understand the basis for this difference, we assessed by qRT-PCR for possible alterations in gene expression profiles for freshly isolated HSCs from matched WT and Dgat1–/– mice. Expression of mRNA was measured for the lipid droplet-related PAT protein genes (Plin1, Plin2, Plin3, Plin4 and Plin5), for genes involved in retinoid metabolism and actions (Lrat, Rbp1, Rarb, Rdh10, Dhrs3, Raldh1 and Cyp26b1), and genes encoding transcription factors that regulate lipid metabolism (Ppara, Ppard, and Pparg, and Srebp1c). The results from this survey are provided in Table 1. We observed no significant differences in expression levels between WT and Dgat1–/– HSCs either for genes associated with lipid droplet formation or for genes involved in the transcriptional regulation of lipid metabolism. Only one of the genes associated with retinoid metabolism and actions was significantly different between the 2 genotypes. Expression of Rbp1 mRNA was significantly elevated (P<0.05) by 2.8-fold in Dgat1–/– HSCs compared to HSCs from WT mice.
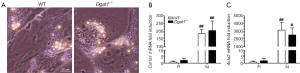
Dgat1–/– HSCs are not protected against activation by culture on plastic dishes
Yamaguchi et al. reported that DGAT1 has a role in HSC activation, reporting that the downregulation of DGAT1 in primary WT HSCs following treatment with antisense oligonucleotides reduced expression levels of genes associated with fibrogenesis (30). We were unable to reproduce this finding in primary HSCs isolated from Dgat1–/– mice. After 5 days of culture, there were no morphological differences in activated Dgat1–/– HSCs compared with WT. The extent of lipid droplet dissolution and dispersal in Dgat1–/– HSCs was visually indistinguishable from that in WT HSCs (Figure 3A). Markers for HSC activation also show that Dgat1–/– HSCs were not less susceptible to activation in culture; Col1a1 and Acta2 mRNA levels were not different in either freshly isolated or cultured HSCs (Figure 3B,C). Cellular retinyl ester levels were not different upon 5 days of culture (data not shown), and mRNA levels for retinoid related genes provide no evidence for elevated retinoic acid or increased retinoic acid signaling in Dgat1–/– HSCs (Table 1). Expression levels of genes involved in the interconversion of retinol and retinaldehyde, Rdh10 and Dhrs3, and in retinoic acid synthesis, Raldh1, as well as the retinoic acid responsive genes, Lrat, Cyp26b1 and Rarb, were all not different between the genotypes for either freshly isolated or activated HSCs (Table 1). However, expression levels of the retinoid-related genes Lrat, Rbp1, Rarb, Rdh10, Dhrs3, and Ralh1 were all significantly elevated in the activated HSCs as compared to freshly isolated HSCs. Similarly, expression levels for Plin2, Plin3 and Plin4 were significantly elevated in culture activated HSCs, although the magnitude of these elevations was identical for WT and Dgat1–/– HSCs. Only expression of Ppard was different (elevated) in the culture activated HSCs but the same degree of elevation was observed for both WT and Dgat1–/– HSCs.
Dgat1–/– livers are protected against CCl4-induced hepatic fibrosis
Although Dgat1-deficiency does not influence culture-induced HSC activation, DGAT1-deficiency may have an influence on HSC activation in situ within the liver. Hence, we investigated the susceptibility of the Dgat1–/– liver to CCl4-induced fibrosis in two studies, which differed by dose and duration of treatment. In Study 1, Dgat1–/– and matching WT mice were given IP injections of 1 µL CCl4 per gram of body weight dissolved in corn oil (1:4, v/v) or an equal volume of the corn oil vehicle every 48 h, for a total of four injections, and were euthanized 24 h after the last injection. Trichrome staining of the liver samples shows that Dgat1-deficiency indeed protects against CCl4-induced fibrosis; the amount of staining clearly reveals less production of extracellular matrix and less inflammatory cell infiltration in the Dgat1–/– liver (Figure 4A). Consistent with this histological analysis, mRNA expression of markers of fibrogenesis, Col1a1 and Acta2, were not different in vehicle-injected livers, but were significantly lower in Dgat1–/– livers when exposed to CCl4 (Figure 4B,C). Following CCl4 treatment, the Dgat1–/– livers had significantly higher retinyl ester levels compared to WT livers (Figure 4D). Compared to vehicle-treated livers, hepatic retinyl ester levels were significantly lower in WT livers after CCl4 treatment; however, CCl4 treatment did not significantly alter retinyl ester levels in the Dgat1–/– livers (Figure 4D).
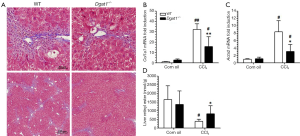
Despite these differences in fibrogenesis, there was no indication of elevated retinoic acid signaling in the Dgat1–/– liver. For both genotypes, CCl4 exposure did not alter expression levels of the retinoic acid-responsive Lrat and Rarb genes (Table 2). Expression levels of Rbp1 were elevated in both WT and Dgat1–/– CCl4-treated livers. However, mRNA levels of Rdh10, Dhrs3, and Raldh1 were all significantly lower for both WT and Dgat1–/– livers compared to vehicle-treated livers (Table 2). CCl4 treatment also significantly lowered Cyp26a1 expression in WT livers, but not in Dgat1–/– livers.
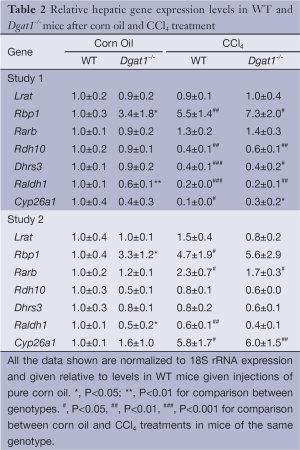
Full table
In Study 2, Dgat1–/– and WT mice were given IP injections of either 0.5 µL of CCl4 per gram of body weight dissolved in corn oil (1:4, v/v) or an equal volume of corn oil vehicle every 7 days, for a total of four injections, and euthanized 7 days after the last injection. Trichrome staining of the liver sections shows a more moderate difference in collagen deposition between the genotypes compared to that in Study 1. As seen in Figure 5A, CCl4 treatment induced more extracellular matrix production and more infiltration of inflammatory cells in the WT liver than in the Dgat1–/– liver. Levels of mRNA of Col1a1 were significantly lower in the CCl4-treated Dgat1–/– liver compared to the CCl4-treated WT liver (Figure 5B). Expression levels of Acta2 mRNA were elevated 2-fold with CCl4 treatment in the WT liver, although this upregulation is modest compared to the 8-fold increase seen in Study 1 (Figure 5C). In the Dgat1–/– liver, Acta2 expression was not statistically different after CCl4 treatment compared to vehicle treatment (Figure 5C). Following CCl4 treatment, retinyl ester levels were significantly higher in Dgat1–/– livers compared to WT livers (Figure 5D).
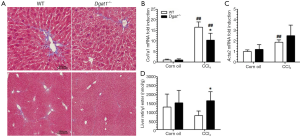
After exposure to CCl4, we observed no differences in hepatic mRNA expression for any of the retinoid related genes between the WT and Dgat1–/– mice (Table 2). For both genotypes, mRNA levels of Lrat, Rdh10 and Dhrs3 were not affected by CCl4 treatment. In contrast to Study 1, hepatic mRNA levels of Rarb and Cyp26a1 were significantly elevated upon CCl4 treatment for both WT and Dgat1–/– mice (Table 2).
Discussion
The goal of our investigations was to answer two questions regarding potential actions of DGAT1 in the liver. First, we wished to understand whether DGAT1, which in some tissues and in vitro catalyzes retinyl ester formation, has a role in retinol/retinyl ester metabolism and storage in the liver. And second, we wanted to understand whether, as proposed by Yamaguchi et al., DGAT1 influences HSC activation through effects on retinoid metabolism and actions (30).
Although we earlier reported that hepatic total retinol (retinol + retinyl ester) levels are not different for age-, gender- and genetic background-matched chow fed WT and Dgat1–/– mice (27,29), in the present study we unexpectedly observed differences in the cellular distribution of retinyl ester stores between hepatocytes and HSCs in the livers of WT and Dgat1–/– mice. While hepatic total retinol levels are not different for WT versus Dgat1–/– liver, agreeing with our earlier work, significantly more retinyl ester is found in Dgat1–/– HSCs compared to WT HSCs, while significantly more retinyl ester is found in WT hepatocytes compared to Dgat1–/– hepatocytes. It is unlikely that this involves the ARAT activity of DGAT1. We were unable to show that DGAT1 catalyzes any retinyl ester formation in the liver, even in the absence of Lrat, or the combined absence of Lrat and Rbp1, or the combined absence of Lrat and Rbp1 combined further with chronic administration of a 25-fold excess of dietary retinol (over that present in a standard chow diet) (27,29). Consequently, there is no basis for attributing this observation to DGAT1-catalyzed retinyl ester formation. It is well established that dietary retinoid is taken up by hepatocytes and then transferred to HSCs for storage (16,17) and that retinol stored in HSCs must be transferred to hepatocytes, the cellular site of retinol-binding protein 4 (RBP4) synthesis within the liver (16), before it can be mobilized in the circulation bound to RBP4. Thus, there are dynamic interactions between these two hepatic cells types that are required for hepatic retinoid uptake, storage and mobilization to occur. We propose that DGAT1 has a role in these processes that is disrupted in Dgat1-deficiency. Rbp1 mRNA levels are elevated by 2.8-fold in HSCs of Dgat1–/– mice (see Table 1). RBP1 is known to have a role in the cellular uptake of retinol and its delivery to LRAT for esterification (39,40). Since RBP1 is highly expressed in HSCs (16,31) and LRAT is almost entirely localized to this cell type within the liver (41), it seems likely to us that the absence of DGAT1 increases the capacity of HSCs for retinyl ester formation and storage. This will disrupt the normal equilibrium in the bidirectional movement of retinol between HSCs and hepatocytes, resulting in increased accrual of retinyl ester by the HSCs. This conclusion is further supported by the data from the studies involving oral administration of [3H]retinol to WT and Dgat1–/– mice (Figure 1E,F). The molecular basis for how Dgat1–/–deficiency gives rise to elevated Rbp1 expression in the HSC will need to be determined in future studies.
The lipid droplets present in HSCs are unique in that they contain large amounts of retinyl ester (approximately 40% of the total lipid) that exceed the concentrations of triglyceride (approximately 33% of total lipid) (42). This is unlike the much more extensively studied lipid droplets that are present in adipocytes whose lipid composition consists predominantly of triglyceride (43). From genetic studies, it has been established that either DGAT1 or DGAT2 can synthesize the triglyceride needed to maintain adipocyte lipid droplets, since only the absence of both Dgat1 and Dgat2 gives rise to the complete absence of lipid droplets in adipocytes (44). DGAT1 is also known to have a role in lipid droplet formation in the hepatocyte (45-47). As an endoplasmic reticulum (ER)-associated enzyme with a dual membrane topology (46,48), DGAT1 is thought to be the DGAT activity responsible for triglyceride synthesis within the ER lumen of hepatocytes (47), and has been proposed to contribute to a population of small lipid droplets, 0.1-0.3 µm in size, destined for the cytosol (49). If these lipid droplets contain retinoids, then the capacity for retinoid accumulation in hepatocytes may also be compromised in the absence of DGAT1, leaving more retinol to be transferred to HSCs for storage.
The lipid droplets present in Dgat1-deficient HSCs are fewer in number but, on average, larger than those of WT HSCs (Figure 2). This observation is consistent with the proposed role of DGAT1 in the formation of small lipid droplets (49). We were not able to attribute these differences in lipid droplet size and number to other parameters measured. We did not observe differences in the HSC levels of triglyceride between WT and Dgat1–/– HSCs (see Figure 1I) indicating that the differences are not simply due to impairments in HSC triglyceride synthesis/accumulation. It is well established in the literature that many different proteins can be associated with lipid droplets and that many of these can affect lipid droplet formation and size (50-52). These include Plin1-5 and enzymes involved in the synthesis and degradation of neutral lipids and phospholipids (50-52). Although we did not carry out an exhaustive study of these, we obtained no evidence that changes in expression levels of the Plin proteins contribute to the differences we observed. Unlike LRAT, which is absolutely required for lipid droplet formation in HSCs (21), DGAT1 appears to have a role in the synthesis of smaller and more numerous HSC lipid droplets (49).
Yamaguchi et al. reported that knockdown of Dgat1 using anti-sense oligonucleotides reduced mRNA expression levels of markers for HSC activation both in mice fed a methionine choline-deficient diet and in cultured, plastic activated HSCs obtained from healthy rats (30). These investigators further reported that treated livers/HSCs displayed elevated Rbp1 and Lrat mRNA expression and concluded that the loss of DGAT1, which they maintained is an important ARAT in liver, caused more retinol to be available for oxidation to retinoic acid, hence suppressing HSC activation (30). Since the only role for DGAT1 in hepatic/HSC retinoid metabolism that can be identified from our studies is an effect on the distribution of retinyl ester in HSCs [present study and (27,29)], we investigated how the complete absence of DGAT1 from the liver affects HSC activation upon culture on plastic dishes and upon CCl4 exposure in vivo, two experimental protocols commonly used to activate HSCs (53). For plastic activation (see Figure 3 and Table 1), the absence of DGAT1 had no effect on HSC activation. Markers of HSC activation were identical for WT and Dgat1-deficient HSCs cultured for 5 days on plastic. Moreover, as noted above, the only difference in gene expression we did observe was for Rbp1, which was elevated by 2.8-fold in freshly isolated Dgat1-deficient HSCs. There were no differences in expression of the canonical retinoic acid-responsive Lrat and Rarb genes. Upon CCl4 treatment, for both study 1 and study 2, and in agreement with the report from Yamaguchi et al., we observed less fibrosis development in the Dgat1–/– mice as assessed both by trichrome staining and by marker gene expression. We did not, however, observe differences between WT and Dgat1–/– livers with regards to expression levels of the retinoic acid-responsive Lrat, Rarb and Cyp26a1 genes. Expression of these genes would be expected to be elevated if retinoic acid was contributing to the lessened fibrosis. Since the literature indicates that different experimental protocols used to activate HSCs (culture on plastic versus CCl4 treatment versus bile duct ligation) each involves different patterns of gene expression (53), it may not be too surprising that we observed different effects for Dgat1-deficiency on fibrogenesis for the culture versus CCl4 experiments. However, we did not obtain evidence from either experiment that supports the notion that differences in retinoic acid synthesis or transcriptional regulatory activity account for the diminished activation observed for Dgat1–/– HSCs.
In summary, our studies establish that DGAT1 has a role in hepatic retinol/retinyl ester metabolism and storage, albeit one that does not involve the ARAT activity of DGAT1. Rather, our data point to a role that involves the movement of retinol between hepatocytes and HSCs, probably directly involving the actions of RBP1 in facilitating retinol uptake and/or LRAT catalyzed esterification in the HSC. Moreover, DGAT1 has a role in modulating the number and size of lipid droplets in HSCs, which is consistent with its proposed actions on triglyceride incorporation into lipid droplets (49). Finally, Dgat1-deficient livers appear to be more resistant to experimentally induced HSC activation and fibrosis. However, we obtained no evidence that this results from differences in retinoid metabolism or retinoic acid synthesis and actions between WT and Dgat1–/– mice as proposed by Yamaguchi et al. (30).
Acknowledgements
This work was supported by U.S. Public Health Services, National Institutes of Health grants R01 DK068437 and R21 AA021336.
Disclosure: The authors declare no conflict of interest.
Ethical Statement: All experiments were conducted in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals, and were approved by the Columbia University Institutional Animal Care and Use Committee.
References
- Mangelsdorf DJ, Umesono K, Evans RM. The retinoid receptors. In: Sporn MB, Roberts AB, Goodman DS, editors. The Retinoids: Biology, Chemistry, and Medicine, 2nd Edition. New York: Raven Press, 1994:319-49.
- Chambon P. The retinoid signaling pathway: molecular and genetic analyses. Semin Cell Biol 1994;5:115-25. [PubMed]
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res 2002;43:1773-808. [PubMed]
- Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res 2013;54:1761-75. [PubMed]
- Rochette-Egly C. Retinoic acid signaling and mouse embryonic stem cell differentiation: Cross talk between genomic and non-genomic effects of RA. Biochim Biophys Acta 2015;1851:66-75.
- Rhinn M, Dollé P. Retinoic acid signalling during development. Development 2012;139:843-58. [PubMed]
- Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol 2011;6:345-64. [PubMed]
- Yang Q, Graham TE, Mody N, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005;436:356-62. [PubMed]
- Graham TE, Yang Q, Blüher M, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 2006;354:2552-63. [PubMed]
- Klöting N, Graham TE, Berndt J, et al. Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass. Cell Metab 2007;6:79-87. [PubMed]
- Maher JJ. Retinol binding protein 4 and fatty liver: A direct link? Hepatology 2013;58:477-9. [PubMed]
- Xia M, Liu Y, Guo H, et al. Retinol binding protein 4 stimulates hepatic sterol regulatory element-binding protein 1 and increases lipogenesis through the peroxisome proliferator-activated receptor-γ coactivator 1β-dependent pathway. Hepatology 2013;58:564-75. [PubMed]
- Bobbert P, Weithäuser A, Andres J, et al. Increased plasma retinol binding protein 4 levels in patients with inflammatory cardiomyopathy. Eur J Heart Fail 2009;11:1163-8. [PubMed]
- Chavarria N, Kato TS, Khan R, et al. Increased levels of retinol binding protein 4 in patients with advanced heart failure correct after hemodynamic improvement through ventricular assist device placement. Circ J 2012;76:2148-52. [PubMed]
- O'Byrne SM, Blaner WS. Retinoids and the metabolic syndrome. Expert Rev Endocrinol Metab 2008;3:539-41.
- Blaner WS, O'Byrne SM, Wongsiriroj N, et al. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta 2009;1791:467-73.
- Blomhoff R, Green MH, Green JB, et al. Vitamin A metabolism: new perspectives on absorption, transport, and storage. Physiol Rev 1991;71:951-90. [PubMed]
- Kluwe J, Wongsiriroj N, Troeger JS, et al. Absence of hepatic stellate cell retinoid lipid droplets does not enhance hepatic fibrosis but decreases hepatic carcinogenesis. Gut 2011;60:1260-8. [PubMed]
- MacDonald PN, Ong DE. A lecithin:retinol acyltransferase activity in human and rat liver. Biochem Biophys Res Commun 1988;156:157-63. [PubMed]
- Batten ML, Imanishi Y, Maeda T, et al. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem 2004;279:10422-32. [PubMed]
- O'Byrne SM, Wongsiriroj N, Libien J, et al. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT). J Biol Chem 2005;280:35647-57. [PubMed]
- Liu L, Gudas LJ. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J Biol Chem 2005;280:40226-34. [PubMed]
- Rasmussen M, Petersen LB, Norum KR. The activity of acyl CoA: retinol acyltransferase in the rat: variation with vitamin A status. Br J Nutr 1984;51:245-53. [PubMed]
- Yen CL, Stone SJ, Koliwad S, et al. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res 2008;49:2283-301. [PubMed]
- Yen CL, Monetti M, Burri BJ, et al. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J Lipid Res 2005;46:1502-11. [PubMed]
- Orland MD, Anwar K, Cromley D, et al. Acyl coenzyme A dependent retinol esterification by acyl coenzyme A: diacylglycerol acyltransferase 1. Biochim Biophys Acta 2005;1737:76-82.
- Wongsiriroj N, Piantedosi R, Palczewski K, et al. The molecular basis of retinoid absorption: a genetic dissection. J Biol Chem 2008;283:13510-9. [PubMed]
- Shih MY, Kane MA, Zhou P, et al. Retinol Esterification by DGAT1 Is Essential for Retinoid Homeostasis in Murine Skin. J Biol Chem 2009;284:4292-9. [PubMed]
- Wongsiriroj N, Jiang H, Piantedosi R, et al. Genetic dissection of retinoid esterification and accumulation in the liver and adipose tissue. J Lipid Res 2014;55:104-14. [PubMed]
- Yamaguchi K, Yang L, McCall S, et al. Diacylglycerol acyltranferase 1 anti-sense oligonucleotides reduce hepatic fibrosis in mice with nonalcoholic steatohepatitis. Hepatology 2008;47:625-35. [PubMed]
- Blaner WS, Hendriks HF, Brouwer A, et al. Retinoids, retinoid-binding proteins, and retinyl palmitate hydrolase distributions in different types of rat liver cells. J Lipid Res 1985;26:1241-51. [PubMed]
- Blaner WS, Dixon JL, Moriwaki H, et al. Studies on the in vivo transfer of retinoids from parenchymal to stellate cells in rat liver. Eur J Biochem 1987;164:301-7. [PubMed]
- Yamada M, Blaner WS, Soprano DR, et al. Biochemical characteristics of isolated rat liver stellate cells. Hepatology 1987;7:1224-9. [PubMed]
- Liu L, Yu S, Khan RS, et al. DGAT1 deficiency decreases PPAR expression and does not lead to lipotoxicity in cardiac and skeletal muscle. J Lipid Res 2011;52:732-44. [PubMed]
- Sohlenius-Sternbeck AK. Determination of the hepatocellularity number for human, dog, rabbit, rat and mouse livers from protein concentration measurements. Toxicol In Vitro 2006;20:1582-6. [PubMed]
- Blouin A, Bolender RP, Weibel ER. Distribution of organelles and membranes between hepatocytes and nonhepatocytes in the rat liver parenchyma. A stereological study. J Cell Biol 1977;72:441-55. [PubMed]
- Pertoft H, Smedsrød B. Separation and characterization of liver cells. In: Pretlow TG II, Pretlow TP, editors. Cell Separation: Methods and Selected Applications Vol. 4. New York: Academic Press, 1987:1-24.
- Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis 2001;21:311-35. [PubMed]
- Ong DE, Newcomer ME, Chytil F. Cellular retinoid-binding proteins. In: Sporn MB, Roberts AB, Goodman DS. eds. The Retinoids: Biology, Chemistry, and Medicine 2nd ed. New York: Raven Press, 1994:283-317.
- Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J 2000;348:481-95. [PubMed]
- Mederacke I, Hsu CC, Troeger JS, et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 2013;4:2823. [PubMed]
- Moriwaki H, Blaner WS, Piantedosi R, et al. Effects of dietary retinoid and triglyceride on the lipid composition of rat liver stellate cells and stellate cell lipid droplets. J Lipid Res 1988;29:1523-34. [PubMed]
- Konige M, Wang H, Sztalryd C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim Biophys Acta 2014;1842:393-401.
- Harris CA, Haas JT, Streeper RS, et al. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J Lipid Res 2011;52:657-67. [PubMed]
- Zammit VA. Hepatic triacylglycerol synthesis and secretion: DGAT2 as the link between glycaemia and triglyceridaemia. Biochem J 2013;451:1-12. [PubMed]
- Wurie HR, Buckett L, Zammit VA. Evidence that diacylglycerol acyltransferase 1 (DGAT1) has dual membrane topology in the endoplasmic reticulum of HepG2 cells. J Biol Chem 2011;286:36238-47. [PubMed]
- Wurie HR, Buckett L, Zammit VA. Diacylglycerol acyltransferase 2 acts upstream of diacylglycerol acyltransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. FEBS J 2012;279:3033-47. [PubMed]
- McFie PJ, Stone SL, Banman SL, et al. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the n terminus in dimer/tetramer formation. J Biol Chem 2010;285:37377-87. [PubMed]
- Thiam AR, Farese RV Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 2013;14:775-86. [PubMed]
- Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 2007;48:2547-59. [PubMed]
- Miyoshi H, Perfield JW 2nd, Obin MS, et al. Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. J Cell Biochem 2008;105:1430-6. [PubMed]
- Ducharme NA, Bickel PE. Lipid droplets in lipogenesis and lipolysis. Endocrinology 2008;149:942-9. [PubMed]
- De Minicis S, Seki E, Uchinami H, et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 2007;132:1937-46. [PubMed]


