
Impact and clinical usefulness of genetic data in the surgical management of colorectal cancer liver metastasis: a narrative review
Introduction
There has been a recent revolution in the availability and analysis of genetic data in resectable colorectal cancer liver metastases (CRLM). For example, although only two studies assessed the prognostic value of KRAS mutation status prior to 2000, at least 15 studies have been subsequently published (1). Similarly, all BRAF studies prior to 2018 cumulatively included only 22 patients with BRAF mutations, but in 2018 alone, the three largest studies to date were published, with 99 patients in total (2-4). Finally, in the last couple years, a few studies were published on other somatic mutations (e.g., APC, P53 and SMAD-4) that were never investigated before (5-7).
In terms of biological activity, KRAS and BRAF belong to the same pathway while APC, P53 and SMAD-4 are members of independent signaling pathways. Specifically, KRAS encodes a small GTP‐binding protein that lies upstream of BRAF and transduces signals from activated cell surface receptors to the nucleus. In turn, these signals control cellular proliferation, differentiation, and survival (8). The BRAF gene encodes a protein kinase downstream from RAS in the canonical mitogen‐activated protein kinase pathway (9). The resultant BRAF mutant is constitutively activated, signals independently of RAS activation, and ultimately leads to increased cellular proliferation and survival. APC (adenomatous polyposis coli) is a tumor suppressor gene involved in the Wnt/β-catenin signaling pathway while TP53 is a tumor suppressor gene in the P53 pathway that prevents cells from progressing through the cell cycle (10,11). Lastly, SMAD-4 is a tumor suppressor gene and as an intracellular transmitter of TGF-β signals. It regulates cell proliferation, differentiation, morphogenesis, and apoptosis (12).
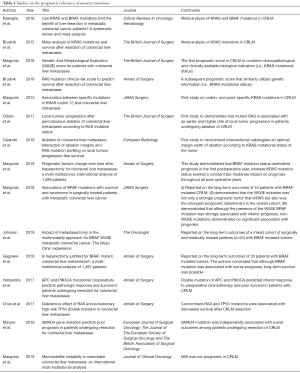
From existing studies (Table 1), we learned that some somatic mutations are consistently prognostic (13-15). Specifically, BRAF and KRAS mutations have been repeatedly associated with worse overall (OS) and recurrence-free survival (RFS). Although useful, an association with prognosis does not necessarily equate to an impact on surgical management, which instead may be influenced (Table 2) by influencing: (I) post-hepatectomy surveillance; (II) selection of patients for surgery; (III) selection of surgical technique; and (IV) selection of optimal margin width. We conducted a computerized search using PubMed and Google Scholar for reports published so far, using mesh headings and keywords related to genetic data and CRLM.
Full table

Full table
We present the following article in accordance with the Narrative Review reporting checklist (available at https://hbsn.amegroups.org/article/view/10.21037/hbsn.2019.10.05/rc).
Discussion
(I) Informing post-hepatectomy surveillance
According to Yaeger et al., patients with KRAS mutated primary colorectal cancers are more likely to develop brain, bone, and lung metastases compared to patients with wild-type tumors (16). In contrast, they are not more likely to metastasize to the liver compared to their wild-type counterparts.
Interestingly, the pattern of recurrence following hepatectomy for CRLM closely resembles the pattern of initial metastatic spread of mutated KRAS tumors. For example, Kemeny et al. found that following hepatectomy for CRLM, patients with KRAS mutated tumors were more likely to develop brain (14.5% vs. 2%, P=0.0533), bone (13.4% vs. 2%, P<0.01), and lung metastases (58% vs. 33.2%, P<0.01) (17). Vauthey and colleagues corroborated that patients with KRAS mutated tumors were more likely to experience lung recurrences compared to patients with wild type tumors (64.7% vs. 38.3%, respectively, P=0.005) (18). Finally, studies from the author’s group and MD Anderson have demonstrated that patients with CRLM and KRAS mutated tumors are not more likely to recur in the liver following CRLM resection (43.8% vs. 50.2%, P=0.181 and 39.0% vs. 52.1%, P=0.1) (18,19).
The prediction of brain and bone recurrences after CRLM resection may not carry clinical significance given their low incidence and the limited amenability of the former to surgical treatment. In contrast, lung recurrences constitute over half of all recurrences in KRAS mutated CRLM and are commonly amenable to surgery. As such, early identification of these recurrences may inform both prognosis and surgical management. Specifically, KRAS status can inform postoperative surveillance, as follow up thoracic imaging can be intensified in patients with KRAS mutated tumors. Furthermore, using codon 13 KRAS mutations instead of the overall KRAS mutation status may improve identification of those at risk for lung recurrence. A study by the author’s group showed that those with tumors harboring codon 13 mutations were at increased risk for lung-specific recurrence compared to other codon-specific KRAS mutations (20). Although it is reasonable to recommend a change in postoperative follow up based on KRAS mutational status, future studies should directly assess whether intensified thoracic follow up in patients with KRAS mutated tumors is associated with improved survival.
(II) Selection of patients for surgery
It has been argued that technical resectability does not equate to biological resectability. This may hold especially true in an era of expanded indications for CRLM surgery, which leads to operations on patients with biologically aggressive tumors. The use of either genetic data alone or in combination with clinicopathologic characteristics may help identify those with biologically aggressive tumors and guide surgical selection. However, even with genetic data, preoperatively identifying patients who will not benefit from surgery has so far proven elusive. The reasons will be discussed below.
Biomarkers alone
It has long been postulated that BRAF mutations confer such a dismal prognosis that resection of CRLM may be futile. In fact, the author’s group recently proved that BRAF mutation was the strongest prognostic factor among all genetic and clinicopathologic factors (2). Although it is reasonable to hypothesize that BRAF may be used to select patients for surgery, we need to first compare survival in surgically vs. medically treated patients with BRAF mutations.
Regarding the former, a multi-institutional, international study led by Johns Hopkins Hospital (JHH) reported on 43 patients with BRAF-mutated CRLM (2). Patients with mutBRAF/wtKRAS tumors had a median OS of 26 compared to 60 months, respectively (P<0.05). Subsequently, Gagnière et al. from Memorial Sloan Kettering (MSK) reported on 35 patients with BRAF-mutated CRLM. Median OS for patients with BRAF mutated and wild-type tumors was 40 vs. 81 months, respectively (P<0.001) (4). Of note, among the 13 patients who were alive at last follow up, there were six 5-year survivors. Lastly, Johnson et al. from Mayo clinic reported on a total of 52 patients. Twenty-one underwent resection and 31 were treated with systemic therapies. Median OS in the surgery vs. medical group was 29.1 vs. 22.7 months (P=0.01) (3).
Collectively, median OS for the surgical cohort ranged between 26–40 months, which is longer than that recently reported for a medical cohort (20 months) (21). This trend may be even more pronounced in reality, as this particular cohort of medically treated patients included both BRAF and wild-type patients. Only the Mayo study compared survival in surgically vs. medically treated BRAF patients, and found that the former had superior survival (3). Although a study that matches medically vs. surgically treated patients is still needed, there is currently no evidence that BRAF mutation alone is a contraindication to surgery.
Biomarkers combined with clinicopathologic data
There are two studies that combined the prognostic powers of both genetic and clinicopathologic data. In the first study, the author’s group collaborated with MSK and constructed the GAME score, in which patients are assigned points according to the presence and prognostic weight of different variables as calculated in the regression analysis (22). The factors were KRAS mutation status (1 point), carcinoembryonic antigen level 20 ng/mL or more (1 point), primary tumor lymph node metastasis (1 point), tumor burden score between 3 and 8 (1 point) or 9 and over (2 points), and extrahepatic disease (2 points). The group with the highest GAME score (6–7 points) had an expected survival rate of 0%. Therefore, it is possible that these patients received no benefit from surgery. However, the number of patients in this group was too small for definite conclusions to be drawn.
A similar score was constructed by the MD Anderson group (23). This time the prognostic weight of each variable was ignored and each of following variables was assigned 1 point: RAS mutation, positive primary tumor lymph node status, and a diameter of the largest liver metastasis >50 mm.
There are 4 major concerns regarding those scores: (I) primary tumor location (PTL), which has recently emerged as a strong prognostic factor, was not included in either of the two scores; (II) the set of clinicopathologic factors largely differed, making their generalizability questionable; (III) scores have not underwent extensive external validations, thus limiting their use outside the cohorts in which they were derived; (IV) neither score had a C index higher than 0.65, which alludes to their limited ability to accurately predict survival. This is unfortunately also true of all previous clinical scores.
Why biomarkers and clinical/genetic scores fail
There are three possible reasons that may explain why existing scores fail to identify patients who will not benefit from surgery. Two of the three may be remedied at present.
Firstly, all existing scores are limited by providing only static survival estimates. The risk of death is calculated only once at the time of surgery under the assumption that the risk of cancer-related death is uniformly distributed over time (actuarial survival). However, evidence indicates that the risk of cancer-related death decreases rapidly over time following surgery. This may explain the empirical observation that some patients with grim prognosis at the time of surgery survive for a long time and refute the initial predictions. If one were to assemble a cohort of such long-term survivors, it is unlikely that the original prognostic factors continue to influence long-term survival. Consequently, factors that were strongly prognostic at baseline may lose their power and new predictors may emerge. The author’s group recently employed a conditional survival analysis to explore this hypothesis in CRLM, and demonstrated that the association of genetic, surgery-related, and clinicopathologic prognostic factors with survival changed dramatically over time (24). Specifically, genetics dominated prognosis in the first year, whereas surgery-related factors (i.e., positive surgical margins and resected extrahepatic disease) determined prognosis thereafter. Although KRAS mutation status was associated with a constant risk of death, the magnitude of its effect was moderate at best. Furthermore, although BRAF is a more important prognosticator, even this mutation may not carry significant prognostic weight a few years following surgery.
Secondly, we need to account for the interplay between different prognostic factors. All current risk score systems assume that variables interact in a linear and additive fashion. Thus, we assign points to each variable according to the hazard ratio calculated in some regression analysis. Those points are added, and the final sum corresponds to a certain risk of death. The mathematical and medical realities, however, suggest that the interactions among prognostic factors are far from linear, and that some variables gain or lose significance due to the absence or presence of other variables (25).
For example, let us consider four variables that have been consistently found to be independent predictors of survival in CRLM: KRAS mutation status, surgical margin status, tumor burden, and primary tumor side. In existing scores, each of these would be treated as “present” or “absent”, and assigned a set number of points irrespective of the presence or absence of the other three factors. Theoretically, however, it may be that patients with KRAS mutated tumors have such a bad prognosis that a positive surgical margin does not further worsen prognosis. Indeed, the author’s group has shown that the impact of margin status differs by KRAS mutation status, as an R0 margin only provided a survival benefit to patients with wild-type tumors (26). Importantly, linear risk score systems would not capture this interplay, and a positive margin may be falsely assigned points in patients with KRAS mutated tumors. As a second example, we can use the interplay between primary tumor laterality and KRAS status. In a recent study, our group found that the prognostic impact of primary tumor laterality differs according to the KRAS mutation status. Right-sided tumors were associated with worse survival only in patients with wild-type tumors. Again, none of the current linear risk score systems capture this interplay.
The interplay between prognostic factors is not limited only to cases where tumor biology is involved. In a third example, patients with high tumor burden theoretically have such a poor prognosis that an R0 margin may not be able to improve survival. This hypothetical scenario has been proved by Oshi et al., who showed that an R0 margin positively impacted prognosis only in patients with low tumor burden (27). In this case, any of the existing linear scores would falsely assign the same number of points to patients with an R1 margin irrespective of tumor burden.
In a non-linear risk score system, prognosis is individualized. For example, the KRAS mutation status would determine whether we include PTL or margin status in the prognostic model for a particular patient. As such, we would individualize not only the weight of each factor, but also which ones to include. Therefore, the set of variables in this non-linear model would be different across patients, and the number of interactions may be so high that only new technologies such as machine learning can address them.
Finally, the third reason cannot be addressed at present. The fact that none of the current risk scores has surpassed a C index of 0.65 alludes to unknown confounders, as it is too low to be attributed solely to the two reasons addressed above. For example, risk scores in other liver malignancies like HCC have a C index of 0.9, despite not accounting for either the interplay of factors or temporal changes in their prognostic power (28). The only remedy is to perform next generation sequencing (NGS) and identify all genetic mutations.
(III) Selection of surgical technique
Currently, there is no evidence to support the use of genetic biomarkers in patient selection for surgery. However, there is evidence to suggest that KRAS mutation status can be used to tailor surgical technique. A study from the author’s group demonstrated that among patients with KRAS mutated tumors, those with anatomical resections (AR) had a RFS at least three times longer than those with non-anatomical ones (NAR) (median RFS: 33.8 vs. 10.5 months, P<0.001) (29). In contrast, among those with KRAS wild-type tumors, surgical technique (AR vs. NAR) was not associated with recurrence rates even on univariable analysis (median RFS: 24.7 vs. 20.4 months, P=0.14). The results were more pronounced for intrahepatic recurrences, suggesting that AR may act as a barrier by removing the major vascular branches that facilitate tumor cell spread to an adjacent liver segment. Of note, OS was not an endpoint of the study. As such, in theory, similar OS could have been achieved if patients underwent repeated hepatectomies for ensuing recurrences instead of a single AR. Even if this was the case, parenchymal sparing hepatectomy could prove to be an inferior strategy in such a scenario, as the same nominal survival gain would be associated with an inferior quality of life due to the need for repeat procedures.
Although there is no similar study to date, a French group indirectly corroborated our results by examining AR vs. NAR, but this time in patients with colorectal cancer lung metastases (30). They found that AR was associated with significantly improved survival and a longer time to pulmonary recurrence in patients harboring KRAS mutations, but not in those with wild-type lung metastases.
Regarding potential shortcomings of the study by our group, in the original study, we included a mixed cohort to reflect “real world” data. However, one might question whether these results could apply to patients who undergo resection alone, given that the study included both patients with resection alone and those who underwent concurrent ablation (31). To address this concern, we also performed a new analysis after excluding the 72 patients with resection and concurrent ablation (32). Importantly, we corroborated our previous findings in the new analysis.
Of note, most surgical series that investigated somatic mutations in CRLM, have included patients treated by surgery and ablation together. The MD Anderson group should be commended on performing the first studies on the effect of RAS mutation on outcomes, after ablation of CRLM. Specifically, they found that mutant RAS was associated with an earlier and higher rate of local tumor progression in patients undergoing ablation alone of CRLM (33). In a subsequent study, the same group extended the practical implications of these findings by demonstrating that an ablation margin of >10 mm, although always desirable, was even more warranted in patients with mutant RAS CRLM to optimize local tumor progression-free survival (34).
(IV) Selection of target margin width
In addition to guiding surgical technique, biomarkers could theoretically inform optimal margin width. There have only been two studies that used a biomarker to recommend an optimal margin width, and the conclusions are inconsistent. Brudvik et al. recommended that surgeons should aim for a 10 mm margin in patients with KRAS mutated tumors (35). In contrast, Margonis et al. found that a 10 mm margin conferred a similar prognosis as a narrow or even positive margin in patients with KRAS mutated tumors (36). In the same study, among those with wild-type tumors, a margin width of 1–4 mm was associated with a survival benefit compared to an R1 resection. Both results are intuitive. It is plausible that a wide margin may be used in an effort to “cancel out” the aggressive biology of a KRAS mutated tumor. On the other hand, it is also plausible that removing a few more millimeters cannot effectively counteract aggressive biology. Regardless of the correct answer, these studies ask the correct question and have triggered a discussion on the interplay between tumor biology and surgical decisions in patients with CRLM.
Refinement of genetic data: from overall mutation status to variants of somatic mutations
Almost all studies have treated mutation status as a binary variable (mutated vs. wild type). However, there is evidence of biological heterogeneity in both KRAS and BRAF somatic mutations. Quoting Haigis, “in the case of KRAS alleles, the devil hides in the details” (8), as different KRAS point mutations activate distinctive downstream signaling pathways (37). This results in varying clinical phenotypes resulting in different degrees of aggressiveness, sites of metastasis, and/or sensitivity to chemotherapy and radiation therapy (38-42). The author’s group was the first to explore the impact of codon- and point-specific KRAS mutations in CRLM (43). Regarding the former, we found that codon 12 but not codon 13 mutations were associated with shorter survival. Regarding the latter, we found that only G12V and G12S mutations were associated with shorter survival. Although the study was limited by small sample sizes in certain sub-groups, the broad message is that labeling patients as mutated vs. wild-type may be limited or even misleading with regards to prognosis. Renaud et al. indirectly corroborated these findings by demonstrating that codon 13 mutations were associated with better outcomes compared to codon 12 mutations in patients who underwent lung metastasectomy for mCRC (38). None of the existing risk score systems has included these sub-groups of KRAS mutations, perhaps because a very large dataset is needed to have sufficient statistical power.
Regarding BRAF mutations, the author’s group recently demonstrated that BRAF mutations in CRLM are also heterogeneous. In a multi-institutional study, we found that only V600E mutations were related to shorter survival and time to recurrence (2). Although this is the first such study in resectable CRLM, studies in unresectable mCRC have reported similar findings. In a study by Cremolini et al., patients with mCRC harboring BRAF codon 594- and 596-mutated tumors outlived both those with wild-type mCRC and BRAF V600E mutations (62 vs. 35.9 vs. 12.6 months) (44). In a subsequent study, Jones et al. corroborated these findings (60.7 vs. 43.0 vs. 11.4 months, respectively; P<0.001) (45). The low incidence of overall BRAF mutations (only 2–4%) and the even lower incidence of certain codon-specific mutations may explain why these mutations have not been included in any clinical/genetic risk scores.
Other less well-studied somatic mutations
In addition to BRAF and KRAS, there are other prognostic somatic mutations such as TP-53, APC, PIK3CA, and MSI, in decreasing frequency. Regarding TP53, Chun et al. recently demonstrated that only a double mutation in RAS/TP53 was prognostic (6). In the same study, a double mutation in APC and PIK3CA was prognostic only in univariable analysis. In contrast, the same group found in another study that a double mutation in APC and PIK3CA was an independent predictor of poor survival (5). These discrepancies may be explained by the small number of patients who had a double APC and PIK3CA mutation (n=45). Lastly, the same group found that SMAD4 mutations were independently associated with worse survival (7). Regarding MSI, the author’s group recently demonstrated that it is not prognostic in CRLM (46). Collectively, these less well studied somatic mutations are either not prognostic or become prognostic only when KRAS is concurrently mutated. As such, they may have limited prognostic impact outside the context of KRAS mutations. Nevertheless, future studies should perhaps assess whether these mutations can tailor surgical margin width/surgical technique as previously shown for KRAS mutations.
Conclusions
In conclusion, genetic data may impact surgical management of CRLM in two ways. Firstly, KRAS mutation status and codon-specific KRAS mutations in particular can predict lung recurrences, and thus inform post-hepatectomy surveillance. Secondly, there is evidence indicating that KRAS mutations may tailor margin width and surgical technique. Nonetheless, whether the use of AR in patients with mutKRAS tumors will prove to be an effective treatment remains to be determined by appropriately designed prospective studies. In contrast, guiding surgical selection remains elusive, as the data cannot support denying surgery to patients according to their KRAS or BRAF mutation profile. Ultimately, we recommend a few axes for future studies. Firstly, future studies should enrich their data with information on mutation sub-groups (KRAS and BRAF codon and point mutations) and integrate them with data on other somatic mutations (P53, PIK3CA, APC). Secondly, the data needs to be analyzed with novel methods, such as machine-learning, that will account for both temporal changes in the prognostic impact and the interplay between different prognostic factors. Thirdly, efforts should be made to uncover unknown genetic data by employing techniques such as next-generation sequencing. Lastly, potential inter- and intra-tumor spatial and temporal heterogeneity might limit the conclusions of current literature and should be taken into account in future studies.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Dr. Timothy Pawlik) for the series “Colorectal Cancer Liver Metastasis” published in Hepatobiliary Surgery and Nutrition. The article was sent for external peer review organized by the editorial office.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://hbsn.amegroups.org/article/view/10.21037/hbsn.2019.10.05/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://hbsn.amegroups.org/article/view/10.21037/hbsn.2019.10.05/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Passiglia F, Bronte G, Bazan V, et al. Can KRAS and BRAF mutations limit the benefit of liver resection in metastatic colorectal cancer patients? A systematic review and meta-analysis. Crit Rev Oncol Hematol 2016;99:150-7. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg 2018;153:e180996. [Crossref] [PubMed]
- Johnson B, Jin Z, Truty MJ, et al. Impact of Metastasectomy in the Multimodality Approach for BRAF V600E Metastatic Colorectal Cancer: The Mayo Clinic Experience. Oncologist 2018;23:128-34. [Crossref] [PubMed]
- Gagnière J, Dupre A, Gholami SS, et al. Is Hepatectomy Justified for BRAF Mutant Colorectal Liver Metastases?: A Multi-institutional Analysis of 1497 Patients. Ann Surg 2020;271:147-54. [Crossref] [PubMed]
- Yamashita S, Chun YS, Kopetz SE, et al. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann Surg 2020;272:1080-85. [Crossref] [PubMed]
- Chun YS, Passot G, Yamashita S, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg 2019;269:917-23. [Crossref] [PubMed]
- Mizuno T, Cloyd JM, Vicente D, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol 2018;44:684-92. [Crossref] [PubMed]
- Haigis KM. KRAS Alleles: The Devil Is in the Detail. Trends Cancer 2017;3:686-97. [Crossref] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006;6:909-23. [Crossref] [PubMed]
- Deming DA, Leystra AA, Nettekoven L, et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 2014;33:2245-54. [Crossref] [PubMed]
- Massagué J. TGFbeta signalling in context. Nat Rev Mol Cell Biol 2012;13:616-30. [Crossref] [PubMed]
- Brudvik KW, Kopetz SE, Li L, et al. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg 2015;102:1175-83. [Crossref] [PubMed]
- Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137-44. [Crossref] [PubMed]
- Yamashita S, Chun YS, Kopetz SE, et al. Biomarkers in colorectal liver metastases. Br J Surg 2018;105:618-27. [Crossref] [PubMed]
- Yaeger R, Cowell E, Chou JF, et al. RAS mutations affect pattern of metastatic spread and increase propensity for brain metastasis in colorectal cancer. Cancer 2015;121:1195-203. [Crossref] [PubMed]
- Kemeny NE, Chou JF, Capanu M, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer 2014;120:3965-71. [Crossref] [PubMed]
- Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013;258:619-26; discussion 626-7. [Crossref] [PubMed]
- Margonis GA, Spolverato G, Kim Y, et al. Effect of KRAS Mutation on Long-Term Outcomes of Patients Undergoing Hepatic Resection for Colorectal Liver Metastases. Ann Surg Oncol 2015;22:4158-65. [Crossref] [PubMed]
- Margonis GA, Kim Y, Sasaki K, et al. Codon 13 KRAS mutation predicts patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Cancer 2016;122:2698-707. [Crossref] [PubMed]
- Adam R, Kitano Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann Gastroenterol Surg 2019;3:50-6. [Crossref] [PubMed]
- Margonis GA, Sasaki K, Gholami S, et al. Genetic And Morphological Evaluation (GAME) score for patients with colorectal liver metastases. Br J Surg 2018;105:1210-20. [Crossref] [PubMed]
- Brudvik KW, Jones RP, Giuliante F, et al. RAS Mutation Clinical Risk Score to Predict Survival After Resection of Colorectal Liver Metastases. Ann Surg 2019;269:120-6. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Prognostic Factors Change Over Time After Hepatectomy for Colorectal Liver Metastases: A Multi-institutional, International Analysis of 1099 Patients. Ann Surg 2019;269:1129-37. [Crossref] [PubMed]
- Chen JH, Asch SM. Machine Learning and Prediction in Medicine - Beyond the Peak of Inflated Expectations. N Engl J Med 2017;376:2507-9. [Crossref] [PubMed]
- Margonis GA, Sasaki K, Kim Y, et al. Tumor Biology Rather Than Surgical Technique Dictates Prognosis in Colorectal Cancer Liver Metastases. J Gastrointest Surg 2016;20:1821-9. [Crossref] [PubMed]
- Oshi M, Margonis GA, Sawada Y, et al. Higher Tumor Burden Neutralizes Negative Margin Status in Hepatectomy for Colorectal Cancer Liver Metastasis. Ann Surg Oncol 2019;26:593-603. [Crossref] [PubMed]
- Halazun KJ, Najjar M, Abdelmessih RM, et al. Recurrence After Liver Transplantation for Hepatocellular Carcinoma: A New MORAL to the Story. Ann Surg 2017;265:557-64. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Anatomical Resections Improve Disease-free Survival in Patients With KRAS-mutated Colorectal Liver Metastases. Ann Surg 2017;266:641-9. [Crossref] [PubMed]
- Renaud S, Seitlinger J, Lawati YA, et al. Anatomical Resections Improve Survival Following Lung Metastasectomy of Colorectal Cancer Harboring KRAS Mutations. Ann Surg 2019;270:1170-7. [Crossref] [PubMed]
- Brudvik KW, Vauthey JN. Surgery: KRAS mutations and hepatic recurrence after treatment of colorectal liver metastases. Nat Rev Gastroenterol Hepatol 2017;14:638-9. [Crossref] [PubMed]
- Margonis GA, Andreatos N, Wolfgang CL, et al. Reply to: "Precision Surgery" for Colorectal Liver Metastases: is the Time Ripe? Ann Surg 2018. [Epub ahead of print]. [Crossref]
- Odisio BC, Yamashita S, Huang SY, et al. Local tumour progression after percutaneous ablation of colorectal liver metastases according to RAS mutation status. Br J Surg 2017;104:760-8. [Crossref] [PubMed]
- Calandri M, Yamashita S, Gazzera C, et al. Ablation of colorectal liver metastasis: Interaction of ablation margins and RAS mutation profiling on local tumour progression-free survival. Eur Radiol 2018;28:2727-34. [Crossref] [PubMed]
- Brudvik KW, Mise Y, Chung MH, et al. RAS Mutation Predicts Positive Resection Margins and Narrower Resection Margins in Patients Undergoing Resection of Colorectal Liver Metastases. Ann Surg Oncol 2016;23:2635-43. [Crossref] [PubMed]
- Margonis GA, Sasaki K, Andreatos N, et al. KRAS Mutation Status Dictates Optimal Surgical Margin Width in Patients Undergoing Resection of Colorectal Liver Metastases. Ann Surg Oncol 2017;24:264-71. [Crossref] [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [Crossref] [PubMed]
- Renaud S, Guerrera F, Seitlinger J, et al. KRAS exon 2 codon 13 mutation is associated with a better prognosis than codon 12 mutation following lung metastasectomy in colorectal cancer. Oncotarget 2017;8:2514-24. [Crossref] [PubMed]
- Renaud S, Schaeffer M, Voegeli AC, et al. Impact of EGFR mutations and KRAS amino acid substitution on the response to radiotherapy for brain metastasis of non-small-cell lung cancer. Future Oncol 2016;12:59-70. [Crossref] [PubMed]
- Renaud S, Seitlinger J, Falcoz PE, et al. Specific KRAS amino acid substitutions and EGFR mutations predict site-specific recurrence and metastasis following non-small-cell lung cancer surgery. Br J Cancer 2016;115:346-53. [Crossref] [PubMed]
- Garassino MC, Marabese M, Rusconi P, et al. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol 2011;22:235-7. [Crossref] [PubMed]
- Renaud S, Falcoz PE, Schaeffer M, et al. Prognostic value of the KRAS G12V mutation in 841 surgically resected Caucasian lung adenocarcinoma cases. Br J Cancer 2015;113:1206-15. [Crossref] [PubMed]
- Margonis GA, Kim Y, Spolverato G, et al. Association Between Specific Mutations in KRAS Codon 12 and Colorectal Liver Metastasis. JAMA Surg 2015;150:722-9. [Crossref] [PubMed]
- Cremolini C, Di Bartolomeo M, Amatu A, et al. BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis. Ann Oncol 2015;26:2092-7. [Crossref] [PubMed]
- Jones JC, Renfro LA, Al-Shamsi HO, et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol 2017;35:2624-30. [Crossref] [PubMed]
- Margonis GA, Buettner S, Wagner D, et al. Microsatellite instability in resectable colorectal liver metastasis: An international multi-institutional analysis. J Clin Oncol 2018;36:220. [Crossref]


