Hepatitis B virus and host factors
Primary liver cancer is the third most common cause of cancer-related deaths worldwide, with a particularly high incidence in Asian countries. Hepatocellular carcinoma (HCC) accounts for 85-90% of primary liver cancers, and is responsible for approximately 660,000 deaths worldwide each year (1). Hepatitis B virus (HBV) is the most common cause of infectious liver diseases. About 400 million people suffer from chronic HBV infection worldwide, and approximately 60% of HCC cases each year are related to chronic HBV infection (2). It has been suggested that viral factors (e.g., HBV-DNA levels, genotypes, and genomic mutations), host factors (e.g., age, sex, race, and immune status), and unhealthy lifestyles might contribute to the progression of liver diseases (3).
Hepatocarcinogenesis is a multi-factorial process in which genetic and epigenetic factors contribute to abnormal activation or inactivation of multiple cellular signaling pathways that result in malignant transformation (4). Integration of HBV into the host’s genome and transactivation of HBx protein promotes and enhances multiple cellular signaling genes (5). For example, this process activates oncogenic genes (e.g., c-myc and c-fos), inactivates suppressor genes (e.g., p53), activates transcriptional factors (e.g., AP-1 and NF-κB), and induces the loss of heterozygosity.
Recent technological advances have revealed that several genetic factors are associated with cancers. Genome-wide association studies (GWAS) have identified numerous single nucleotide polymorphisms (SNPs) that are associated with various cancers. With respect to liver cancers, SNPs in numerous candidate genes were reported to be associated with HCC in case-control and retrospective studies. Table 1 summarizes the results of GWAS identifying genetic factors implicated in HCC. In 2009, Kamatani et al. first reported that SNPs in the human leukocyte antigen (HLA)-DP region were associated with chronic HBV in a study of 188 Japanese patients with chronic HBV infection and 934 controls (6). The HLA gene is located in the region 6p21.3 and plays an important role in antigen presentation. Polymorphisms in this region were also reported in Chinese patients (8,12). In 2010, Zhang et al. reported that SNPs in several tumor suppressor genes located in the region 1p36.22, including KIF1B, UBE4B, and PGD, were putatively associated with HCC (7). However, no data from other countries in Asia, or globally, have been published to confirm these associations. GWAS may result in some discrepancies because of differences among ethnicities. Most of the studies published to date were conducted in Asian countries, particularly China and Japan (Table 1). Therefore, it is necessary to confirm these associations in other countries and ethnicities.
It is also important that such studies include samples from patients and controls. Several studies identified candidate SNPs by comparing the SNPs present in asymptomatic HBV carriers and HCC patients. However, other factors, including age, sex, viral load, and viral mutations, are often clinically very different between these two groups, and may confound the analyses. Therefore, to precisely analyze genetic factors, it is essential that the patients and controls are well matched for these factors to limit possible confounding. Several studies have also enrolled patients with chronic HBV infection and controls without HBV infection. The results of these studies imply that several SNPs are associated with persistent infection and HBV clearance. Functional analyses are necessary to confirm these results.
Chronic hepatitis B (CHB) normally progresses through five phases: immune tolerance, immune clearance, inactive carrier, reactivation, and recovery. However, the duration and underlying hepatitis disease activity vary considerably between each phase. Additionally, the clinical course differs between patients. HBV itself is not usually cytopathogenic. Instead, the liver injuries in patients with chronic HBV infection are considered to be the result of the host’s immune responses against HBV. For example, an HLA-class I antigen-restricted, cytotoxic T lymphocyte-mediated response to the HBV antigen expressed on hepatocytes results in apoptosis and necrosis (20). Viral and host factors are associated with viral clearance, persistent infection, and clinical course. It was reported that HLA-DP variants were associated with chronic persistent hepatitis following HBV infection. The HLA-DP molecule plays an important role in antigen presentation to cytotoxic T cells. Therefore, functional differences in HLA-DP might contribute to differences in host sensitivity to the virus.
Li et al. identified a susceptibility gene within the HLA region, and the SNP identified in their study may influence HLA function. GWAS provide abundant information on a variety of diseases (18). However, the generalizability of the results should still be discussed because the data may differ according to ethnicity and other confounding factors. The pathogenesis of HCC involves multifactorial processes. Further studies are needed to determine whether the susceptibility gene reported by Li et al. shows clinical generalizability, and to determine the importance of this gene in the progression of HCC.
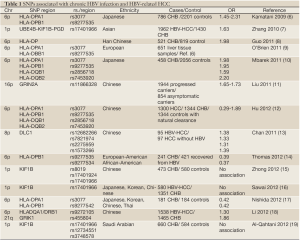
Full table
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76.
- Lai CL, Ratziu V, Yuen MF, et al. Viral hepatitis B. Lancet 2003;362:2089-94.
- Liaw YF. Natural history of chronic hepatitis B virus infection and long-term outcome under treatment. Liver Int 2009;29:100-7.
- Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin Liver Dis 2010;30:3-16.
- Paterlini-Bréchot P, Saigo K, Murakami Y, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene 2003;22:3911-6.
- Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009;41:591-5.
- Zhang H, Zhai Y, Hu Z, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 2010;42:755-8.
- Guo X, Zhang Y, Li J, et al. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 2011;53:422-8.
- O’Brien TR, Kohaar I, Pfeiffer RM, et al. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 2011;12:428-33.
- Mbarek H, Ochi H, Urabe Y, et al. A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 2011;20:3884-92.
- Liu L, Li J, Yao J, et al. A genome-wide association study with DNA pooling identifies the variant rs11866328 in the GRIN2A gene that affects disease progression of chronic HBV infection. Viral Immunol 2011;24:397-402.
- Hu L, Zhai X, Liu J, et al. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 2012;55:1426-31.
- Chan KY, Wong CM, Kwan JS, et al. Genome-wide association study of hepatocellular carcinoma in Southern Chinese patients with chronic hepatitis B virus infection. PLoS One 2011;6:e28798.
- Thomas R, Thio CL, Apps R, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol 2012;86:6979-85.
- Zhong R, Tian Y, Liu L, et al. HBV-related hepatocellular carcinoma susceptibility gene KIF1B is not associated with development of chronic hepatitis B. PLoS One 2012;7:e28839.
- Sawai H, Nishida N, Mbarek H, et al. No association for Chinese HBV-related hepatocellular carcinoma susceptibility SNP in other East Asian populations. BMC Med Genet 2012;13:47.
- Nishida N, Sawai H, Matsuura K, et al. Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS One 2012;7:e39175.
- Li S, Qian J, Yang Y, et al. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet 2012;8:e1002791.
- Al-Qahtani A, Al-Anazi M, Viswan NA, et al. Role of Single Nucleotide Polymorphisms of KIF1B Gene in HBV-Associated Viral Hepatitis. PLoS One 2012;7:e45128.
- Liaw YF. Hepatitis flares and hepatitis B e antigen seroconversion: implication in anti-hepatitis B virus therapy. J Gastroenterol Hepatol 2003;18:246-52.


