PNPLA3 genetic variation in alcoholic steatosis and liver disease progression
Introduction
According to the World Health Organisation, morbidity attributable to heavy alcohol drinking in developed countries accounts for 10.3% of disability adjusted life years and comes second only to that of tobacco (11.7%) (1). The enormous disease burden has an economic impact of about 125 billion Euros annually in Europe, accounting for 1.3% of the gross domestic product. Alcoholic liver cirrhosis is responsible for 70-80% of the directly recorded mortality from alcohol (2).
Alcoholic liver disease (ALD) comprises a continuum of partly overlapping liver lesions with steatosis in nearly all drinkers, necroinflammation and progressive fibrosis in 10-35% of alcoholics, and liver cirrhosis in approximately 10-15% of heavy drinkers (3). Thus, only a minority of drinkers develop severe liver disease suggesting disease modifiers which determine an individual’s risk to progress to significant liver disease when drinking. Several lines of evidence suggest that—besides the patterns and amount of alcohol consumption, liver-related comorbidities and obesity—genetic factors contribute significantly to the liver phenotype in drinkers: (I) women are more susceptible for ALD than men when exposed to similar amounts of alcohol (4); (II) Hispanics are more prone to developing ALD than Blacks and Whites (5); (III) twin studies demonstrate that monozygotic twins have a higher prevalence of alcoholic cirrhosis than dizygotic twins (6,7). In the past 30 years, these observations and the emerging availability and declining costs of genoytping have precipitated an avalanche of genetic studies attempting to identify genetic factors associated with ALD particularly cirrhosis (8). While most putative associations did not pass proper scrutiny or independent replication, a genetic polymorphism in the gene coding for patatin-like phospholipase domain-containing 3 (PNPLA3; adiponutrin; rs738409 C/G, M148I) was first found to associate with hepatic fat content in patients with non-alcoholic fatty liver disease (NAFLD) by means of a genome-wide case control study (9), and subsequently also identified as a risk factor for alcoholic cirrhosis and lesser degrees of ALD (10). Since then, additional genetic studies in humans have been published on the same issue, and experimental insight has been generated to elucidate the biological implications of PNPLA3 function and genetic variation thereof.
The present review aims to summarize the essential knowledge published so far, and to draw possible lines of future research to make use of the revelations made on this important host genetic modifier of alcoholic liver injury.
PNPLA3 variation: genetic studies in human alcoholic
After the initial two genetic case control studies on an association of the single nucleotide polymorphism PNPLA3 rs738409 with hepatic fat content in patients with NAFLD, and with ALD, several candidate gene case control studies investigating PNPLA3 rs738409 were published, yielding similar conclusions and confirming this association. Altogether, ten case control studies have been published with a total of 3,495 patients with ALD of which 2,087 had cirrhosis, compared to 5,038 controls of which 4,007 were healthy subjects and 1,031 active alcoholics without evidence of ALD. Major study details and calculated odds ratios (OR) are listed in Table 1.
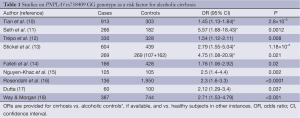
Full table
In the first study in ALD, Tian et al. investigated a possible relationship between PNPLA3 variation and ALD using 17 variants within PNPLA3 and a panel of 306 ancestry-informative SNPs in a mixed European and Native American (Mestizo) population with a history of alcoholism (10). A significiant association of PNPLA3 rs738409 GG with cirrhosis (OR =2.25, P=1.7×10−10) was detected which remained significant after correcting for ancestry (adjusted OR =1.45, P=2.8×10−3).
Independent confirmation came from a small study from the UK and Australia in which 266 alcoholic cirrhotics and 182 heavy drinkers with normal liver laboratory and no clinical evidence of significant liver damage were included (11). Here, carriership of the PNPLA3 rs738409 G allele and GG genotype were also significantly associated with alcoholic cirrhosis with similar OR (2.2 and 5.57, respectively). Although no information on a possible association of PNPLA3 rs738409 with liver enzyme levels or Child-Pugh scores was reported, this study selected appropriate drinking controls rather than healthy subjects thereby avoiding a pitfall of many previous candidate gene case control studies. A similar case control study from Belgium included patients with ALD and healthy non-drinking controls (12). Among ALD patients, 265 had cirrhosis and 65 had lesser degrees of liver injury. Again, the PNPLA3 rs738409 G allele was significantly more frequent in ALD patients than in controls (OR =1.54), and PNPLA3 rs738409 G was found as the strongest independent factor associated with risk of cirrhosis after multivariate analysis (OR =2.08). Interestingly, hepatic PNPLA3 mRNA expression was significantly lower in patients with more advanced fibrosis and negatively correlated with the hepatic venous pressure gradient as a surrogate for portal pressure.
A relatively large study on PNPLA3 variation in alcoholic Caucasians from Germany included 1,043 alcoholic cases and controls from different German hepatology and addiction centres, and an additional population-based cohort for independent validation of association (13). The PNPLA3 rs738409 G allele was most frequent in alcoholic cirrhotics and lowest in drinkers with steatosis and normal serum liver enzyme levels (35% vs. 17.7%). The association of PNPLA3 rs738409 with ALD was confirmed in the independent population-based cohort of drinkers at-risk for ALD, and the population-attributable risk from PNPLA3 rs738409 G was calculated at 26.6%.
Falleti et al. studied 483 cirrhotic patients from Italy of whom 166 had alcoholic cirrhosis (14). The PNPLA3 rs738409 G allele was more frequent in cirrhotics with alcoholic and metabolic etiology, and particularly aggregated in patients with hepatocellular carcinoma (HCC) with an overall OR of 1.76 [95% confidence interval (CI), 1.06-2.92]. Of note, this study was the first one pointing to a possible role of PNPLA3 variation in the risk of HCC. Another study still only available as an abstract addressed the question whether there is a role of PNPLA3 in alcoholic hepatitis (15). Patients with alcoholic cirrhosis and healthy subjects were used as controls compared to cases who were patients with severe alcoholic hepatitis defined by the Maddrey’s discriminant function threshold of >32. The frequency of PNPLA3 rs738409 G allele was not significantly different between cirrhotics and those with complicating alcoholic hepatitis, but markedly more frequent than in healthy controls. However, an intriguing finding of that study was that PNPLA3 rs738409 G allele carriers were more likely to die of alcoholic hepatitis that those with the C allele, providing an example on how genetic findings may be translated into important clinical settings. A large German/Dutch study in 961 patients with alcoholic chronic pancreatitis and 135 patients with alcoholic cirrhosis confirmed the overrepresentation of the PNPLA3 rs738409 G allele in cirrhotics, but ruled out an association with alcoholic pancreatitis, fitting to the organ-specificity of PNPLA3 expression restricted to liver and adipose tissue (16). The only study in Asians stems from India published as a single author paper (17). Dutta studied as small set of 100 healthy controls and 120 alcoholics (60 alcoholic noncirrhotics and 60 alcoholic cirrhotics) by genotyping for ten SNPs previously reported to be associated with ALD in various populations, including PNPLA3. Among these, only PNPLA3 rs738409 was associated with alcoholic cirrhosis with borderline significance.
Way and Morgan recruited 387 heavy drinkers with a history of alcohol abuse of >25 years and liver biopsies showing cirrhosis in 206, lower stages of fibrosis in 114, and 67 who had only mild steatosis (18). In addition, 744 healthy controls were also genotyped for PNPLA3 rs738409 to find the GG genotype associated with cirrhosis with an OR of 2.71 (95% CI, 1.53-4.79). A similar finding was obtained when comparing all alcoholic patients with healthy controls (OR =1.67; 95% CI, 1.00-2.79). An interesting additional finding in that study fitting to the observation made in the erlier French study (15) was that carriers of the PNPLA3 rs738409 G allele had a poorer intermediate term survival.
Taken together, evidence for an eminent role of PNPLA3 rs738409 in determining the risk of alcoholic cirrhosis is striking, and it seems no surprise to see this confirmed in a recent metaanalysis (19).
PNPLA3 variation and HCC
Alcoholics with established cirrhosis who continue to drink not only face the likely fate of declining liver function and subsequent decompensation, but also a significant risk of developing HCC (20). Even in non-drinking alcoholic cirrhotics, HCC has an annual incidence of approximately 1-2% (21). Therefore, current management guidelines suggest implementation of screening of alcoholic cirrhotic patients by ultrasound every 6-months to detect HCC early to allow for curative resection (22,23). Obviously, screening efforts and detection rates could be optimized with markers which allow for focusing on groups particularly at risk. In this regard, PNPLA3 rs738409 could become an interesting biomarker.
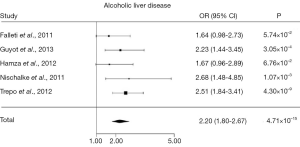
Several genetic case control studies have addressed this issue lately, and a recent meta-analysis has amalgamated the conclusions (24). Of all genetic case control studies investigating PNPLA3 rs738409 in HCC, five studies (14,25-28) included alcoholic cirrhotics with (cases; n=442) and without HCC (control; n=932), showing together that the PNPLA3 rs738409 G allele is associated with HCC in alcoholic cirrhotics with an OR of 2.20 (95% CI, 1.80-2.67, P=4.71×10–15) (Figure 1). The association remained robust after correction for the potential confounders age, gender and body mass index. Interestingly, the association of PNPLA3 rs738409 G allele and GG genotype with HCC was particularly strong in alcoholics, and less evident, but still significant in chronic hepatitis C cirrhosis.
PNPLA3 function
Understanding the physiological function of PNPLA3 and the effect of the genetic variant encoding for the isoleucine to methionine substitution at position 148 of the protein (I148M) has been a challenging task for biomedical scientists after the genetic association was robustly established. With clear and unequivocal genetic associations of the PNPLA3 I148M genetic variant found with diverse liver disease phenotypes including hepatic steatosis (29), necroinflammation (30,31), fibrosis (32,33) and cancer (34,35), gaining a deeper insight to its function and the functional alterations along with its genetic variation was pivotal. In vitro studies using purified PNPLA3 protein (36-38) and cell culture models (39-41) and in vivo studies (42-46) using genetically modified murine models have been performed to understand the molecular basis underlying the genetic association. The main questions on this protein regarding the liver are: where is PNPLA3 localized inside cells and which liver cells express it under which conditions? What is PNPLA3’s enzymatic activity? In which metabolic pathways is PNPLA3 involved and what substrates are metabolized? And finally, what functional change derives from the PNPLA3 I148M variant on these features?
The major findings and the still open questions regarding all these aspects are presented from the simplest models using purified proteins to the most complex animal models. PNPLA3 protein has been related so far to fatty acids metabolism, i.e., triacylglycerol hydrolysis. Therefore, in a final section we will separately take in exam the latest findings regarding a new aspect of PNPLA3 in the regulation of retinoid metabolism.
Enzymatic studies using purified human PNPLA3 protein
The human PNPLA3 gene contains a sequence encoding for a patatin-like phospholipase domain (47,48). This domain characterizes the entire patatin-like protein family and in PNPLA3 it contains a serine at position 47 of the amino acidic sequence and an aspartate at position 166 (40). Based on in silico structural modeling these two amino acids constitute a catalytic dyad for phospholipase activity (47). This model is consistent with a lipase activity of the protein. However, the actual structure of PNPLA3 has never been resolved by crystallization or nuclear magnetic resonance spectroscopy. Thus, it is of great interest to test the enzymatic function of PNPLA3 by using the purified protein based on the predicted protein function. Furthermore, the purified protein approach offers a very simple model to test the enzymatic activity of a protein with virtually no confounders. Three different works have demonstrated that the PNPLA3 protein has triacylglycerol lipase activity (36-38), by incubating the purified protein with radiolabeled triolein and subsequently measuring the release of radiolabeled oleic acid. In two of these works, PNPLA3 has been purified using insect cells Sf-9 as expression system (37,38), whereas in one case the yeast Pichia pastoris has been used (36). PNPLA3 has a higher activity on triacylglycerol containing mono-unsaturated fatty acids as side chains as opposed to saturated and poly-unsaturated. Importantly, the three papers show PNPLA3 being mainly localized on cell membranes. This is consistent with the presence of a predicted trans-membrane domain in this protein. In two (36,37) of the three studies the activity of the purified PNPLA3 148M mutant protein has also been investigated. In these studies the mutation was found to drastically reduce the triacylglycerol lipase activity, indicating that the isoleucine to methionine substitution at position 148 of the PNPLA3 protein causes a loss of function. These finding are in line with a possible reduced lipase activity of the 148M PNPLA3 causing liver fat accumulation/trapping, and subsequent lipotoxic liver damage in humans.
In vitro studies using cell lines
The human PNPLA3 gene is expressed mainly in the liver (49); therefore, most of the in vitro studies on PNPLA3 function were performed using immortalized hepatic cell lines. A common approach used by three independent groups (39-41) was based on overexpressing the wild type and 148M mutant PNPLA3 protein in hepatic cell lines and subsequently examining the effect of the overexpression on metabolic features such as lipid accumulation and lipid secretion. In a study using the human hepatoma-7 (HuH-7) cell line authors could show that the adenovirus-mediated acute overexpression of the mutant 148M PNPLA3 induces a 2-fold increase in intracellular triacylglycerol content measured by thin layer chromatography compared to overexpression of the 148I wild type protein (40). This difference remained after treatment with Triacsin C, an inhibitor of triacylglycerol synthesis, indicating that the 148M mutation induces a reduction of the hydrolysis rather than an increase of triacylglycerol synthesis. In the same set of experiments, PNPLA3 localizes on cell membranes and lipid droplets, with no differences between the wild type and the mutant protein. These results indicate that the amino acidic change does not affect the protein binding to the cell membranes. The effect of different fatty acids on PNPLA3 protein stability was examined by using three different fatty acids on HuH-7 cells. Palmitic acid, oleic acid and linoleic acid, show a stabilizing effect on PNPLA3, thus inducing an increase in the protein intracellular content with no effect on mRNA expression. Finally, a glucose-dependent regulation of PNPLA3 was shown in vitro in HuH-7 cells and human hepatocytes (41). Moreover, PNPLA3 mRNA expression was up-regulated by glucose and this effect abolished by silencing the carbohydrate response element-binding protein (ChREBP) with small interfering RNA. Consistently, PNPLA3 promoter contains a sequence that binds ChREBP, as shown by chromatin immunoprecipitation. The same study has also investigated the effect of the 148M mutation on intracellular lipid content and confirmed that the overexpression of mutant PNPLA3 induces accumulation of triglycerides mediated by reduced hydrolysis.
Very recently, HuH-7 cells overexpressing PNPLA3 148I and 148M have been used to perform lipidomic analysis after incubation with radiolabeled oleic acid or glycerol (50). Herein, the wild type but not the mutant protein increases the turnover of oleic acid. This finding is consistent with data from studies on purified proteins that have identified oleic acid as the best substrate for PNPLA3. Furthermore, the 148M mutant protein induces accumulation of triacylglycerols, but not those newly synthesized. This data point out a possible involvement of PNPLA3 in the de novo lipogenesisis and are consistent with findings from a study in humans, in which carbohydrate overfeeding induced in an increase in de novo lipogenesis proportional to increase in liver fat content and circulating triacylglycerols 148I wild type carriers, but not in 148M mutant individuals (51).
A different approach has been used to study the role of PNPLA3 on hepatic very low density lipoprotein (VLDL) secretion (39). VLDLs are lipoprotein particles highly enriched in triacylglycerol and in fasting conditions nearly represent the total amount of circulating triacylglycerols (52). VLDLs are secreted by the liver to make lipid species available for the peripheral tissues (53). Retention of VLDLs in the liver cause increased hepatic fat content (54). A possible involvement of PNPLA3 in VLDL secretion has been tested by using the hepatic cell line McArdle rat hepatoma cells (McA-RH7777). McArdle cells stably overexpressing the wild type or the mutant PNPLA3 have been generated and used to assess the effect of the 148M mutation on intracellular fat content, VLDL secretion and triacylglycerol hydrolysis. The stable overexpression of the 148M mutant PNPLA3 in this model induced increased triacylglycerol content due to a reduced hydrolysis and coupled with reduced VLDL secretion. This finding has been confirmed in humans by measuring hepatic VLDL secretion by injection of stable isotopes across different PNPLA3 genotypes. The role of PNPLA3 on VLDL composition was shown by a study that identified a striking reduction of a specific subgroup of triacylglycerol containing unsaturated fatty acids as side chains in carriers of the 148M mutant allele (55).
In vivo studies using murine models
Several genetically modified murine models were used in vivo to understand the PNPLA3 function. Up to date adenovirus-mediated acute overexpression (40), antisense oligonucleotide-mediated silencing (56), knock out mouse model (43,45), transgenic mouse model (42) and knock in mouse model (46) were studied. The first in vivo approach to study PNPLA3 function was adenovirus mediated-acute PNPLA3 overexpression in mice (40). This set of experiments showed that 148M mutant protein overexpression increases liver fat content compared to the 148I wild type overexpression. This finding is consistent with the human phenotype. The same result is obtained overexpressing non-functional version of the enzyme, the S47A, where the catalytic site at position 47 was abolished. This indicates that the 148M mutation results in a loss of function in vivo.
Transgenic mice overexpressing the sterol regulatory element binding protein 1a, 1c or 2 (SREPBP-1a, SREBP-1c and SREBP2) were used to assess the transcriptional regulation of PNPLA3 (49). In all three genetically modified mouse model PNPLA3 mRNA is up-regulated. Consistently, treatment with an agonist of LXR, an activator of SREBP-1c, induces the same effect in wild type mice but does not in mice lacking SREBP-1c. Chromatin immunoprecipitation has confirmed the presence in the intron 1 of PNPLA3 gene of a SREBP-1c binding site. This finding implicates an important link between PNPLA3 and the insulin, since SREBP-1c is downstream the insulin signaling (56). Another hint of a link between PNPLA3 and insulin signaling has been provided by a study on rats (55), in which the endogenous Pnpla3 has been silenced using antisense oligonucleotides. In these rats, PNPLA3 silencing improves insulin sensitivity, possibly via reduction in hepatic diacylglycerol content and subsequent reduction in hepatic protein kinase Cε activation. A role of PNPLA3 in glucose homeostasis is also suggested by a study on obese subjects (57) in which carriers of the M mutant allele show an increased risk of type 2 diabetes mellitus.
Two independent PNPLA3 knock-out mouse models have been generated to study the effect of the absence of PNPLA3 in vivo (43,45). Surprisingly, neither showed a liver disease phenotype. This may be explained by a different role of the protein in mice or by compensatory mechanism activated during embryogenesis.
A transgenic mouse model resulting in liver-specific chronic overexpression of human wild type or mutant PNPLA3 has been generated (42). This model confirms that overexpression of the mutant PNPLA3 induces liver fat accumulation, especially triacylglycerols containing mono-unsaturated fatty acids. The more recent and most advanced mouse model available for studying PNPLA3 function is a knock-in mouse model that endogenously expresses the mouse 148M mutant pnpla3 protein or the S47A variant (46). Mice expressing the 148M mutant PNPLA3 develop liver steatosis when fed a high sucrose diet. The same happens for mice expressing the S47A variant, indicating that the I148M mutation is a loss of function. Up to date, this mouse model is the one that more closely resembles in vivo the human physiology. Although studies have shown the protein enzymatic function which is responsible for the hepatic lipid accumulation, most questions regarding the molecular mechanism underlying the association with liver inflammation, fibrosis and cancer are still under investigation.
PNPLA3 and retinoid metabolism
Very recently, a new study opened a new insight on PNPLA3 physiological function on retinol storage (58). Retinol belongs to the Vitamin A family (59). Retinol is stored in lipid droplets within hepatic stellate cells as retinyl ester (mainly retinyl palmitate) (60,61). When vitamin A is needed for its physiological functions (62), such as the vision in the retina (as retinaldehyde) and the tuning of more than 500 genes in the liver (as retinoic acid), retinol is released from the ester bound with palmitic acid. The enzyme responsible for the retinyl hydrolase activity in the stellate cells was not known until a recent study identified PNPLA3 as this enzyme.
In particular, experiments on primary human hepatic stellate cells showed that PNPLA3 is highly expressed and synthesized in these cells and that PNPLA3 catalyzes the hydrolysis of retinyl esters. The 148M mutation abolishes this function resulting in the retention of retinyl palmitate intacellularly. In humans the 148M mutation associates with reduced circulating levels of retinol binding protein 4 (RBP4), the main carrier of vitamin A in the blood stream (63).
In relation to chronic liver disease, upon liver injury hepatic stellate cells change their phenotype from quiescent retinol-storing cells to activated myofibroblast-like cells secreting the collagen responsible for liver fibrosis (64). During this phenotype change, hepatic stellate cells lose their retinol content. It is not known if the retinol loss is a cause, a consequence or just a marker of hepatic stellate cell activation and liver damage. Finally, the overall role of PNPLA3 in this activation process remains to be answered.
Conclusion and future perspective
In summary, data on PNPLA3 allow for the conclusion that PNPLA3 rs738409 GG carriers should be considered a genetically defined subpopulation of high risk alcohol-drinking subjects particularly prone to developing alcohol-related liver injury with the potential to progress to cirrhosis and HCC. Genotype PNPLA3 rs738409 GG is the only hereditary factor in the progression of ALD known so far, but there are likely additional factors involved fitting to the complex genetic background of ALD. What is clearly lacking on ALD genetics are data from genome-wide analyses similar to what has been generated for NAFLD and other chronic liver diseases. Systematic scanning of multiple variants would not only help to confirm PNPLA3 rs738409 as an important susceptibility gene of ALD, but could also identify yet unkown genetic variants which influence the natural course of ALD. Obviously, knowing the complete genetic background of ALD may greatly improve screening and counseling of patients who drink.
Functionally, it has become clear that the PNPLA3 protein is an enzyme with lipase activity towards triacylglycerol and retinyl esters. The naturally occurring mutation resulting into an amino acidic change of the isoleucine to methionine at position 148 of the protein determines a loss of function of this protein leading to intracellular retention/remodeling/trapping of lipid species, and subsequently to hepatic steatosis. The mechanism underlying the genetic susceptibility to liver necroinflammation and fibrosis—although well-established by association to disease phenotypes in humans—is still an open question, as well as it remains elusive how PNPLA3 variation impacts functionally on hepatocarcinogenesis besides contributing to lipotoxicity.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Health statistics and information systems, Estimates for 2000–2012. Available online: http://www.who.int/healthinfo/global_burden_disease/estimates/en/index2.html
- Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J Hepatol 2013;59:160-8. [PubMed]
- Teli MR, Day CP, Burt AD, et al. Determinants of progression to cirrhosis or fibrosis in pure alcoholic fatty liver. Lancet 1995;346:987-90. [PubMed]
- Sato N, Lindros KO, Baraona E, et al. Sex difference in alcohol-related organ injury. Alcohol Clin Exp Res 2001;25:40S-45S. [PubMed]
- Stinson FS, Grant BF, Dufour MC. The critical dimension of ethnicity in liver cirrhosis mortality statistics. Alcohol Clin Exp Res 2001;25:1181-7. [PubMed]
- Hrubec Z, Omenn GS. Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcohol Clin Exp Res 1981;5:207-15. [PubMed]
- Reed T, Page WF, Viken RJ, et al. Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res 1996;20:1528-33. [PubMed]
- Stickel F, Hampe J. Genetic determinants of alcoholic liver disease. Gut 2012;61:150-9. [PubMed]
- Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461-5. [PubMed]
- Tian C, Stokowski RP, Kershenobich D, et al. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 2010;42:21-3. [PubMed]
- Seth D, Daly AK, Haber PS, et al. Patatin-like phospholipase domain containing 3: a case in point linking genetic susceptibility for alcoholic and nonalcoholic liver disease. Hepatology 2010;51:1463-5. [PubMed]
- Trépo E, Gustot T, Degré D, et al. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J Hepatol 2011;55:906-12. [PubMed]
- Stickel F, Buch S, Lau K, et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology 2011;53:86-95. [PubMed]
- Falleti E, Fabris C, Cmet S, et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int 2011;31:1137-43. [PubMed]
- Nguyen-Khac E, Houchi H, Dreher ML, et al. Is PNPLA3 polymorphism involved in severe acute alcoholic hepatitis. Hepatology 2011;54:976A.
- Rosendahl J, Tönjes A, Schleinitz D, et al. A common variant of PNPLA3 (p.I148M) is not associated with alcoholic chronic pancreatitis. PLoS One 2012;7:e29433. [PubMed]
- Dutta AK. Genetic factors affecting susceptibility to alcoholic liver disease in an Indian population. Ann Hepatol 2013;12:901-7. [PubMed]
- Way MJ, Morgan M. The PNPLA3 I148m mutation significantly increases the risk of developing alcohol-related cirrhosis in alcohol dependent individuals. Alcohol Alcohol 2013;48:37.
- Chamorro AJ, Torres JL, Mirón-Canelo JA, et al. Systematic review with meta-analysis: the I148M variant of patatin-like phospholipase domain-containing 3 gene (PNPLA3) is significantly associated with alcoholic liver cirrhosis. Aliment Pharmacol Ther 2014;40:571-81. [PubMed]
- Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer 2007;7:599-612. [PubMed]
- Fattovich G, Stroffolini T, Zagni I, et al. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology 2004;127:S35-50. [PubMed]
- European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908-43. [PubMed]
- Sharma P, Saini SD, Kuhn LB, et al. Knowledge of hepatocellular carcinoma screening guidelines and clinical practices among gastroenterologists. Dig Dis Sci 2011;56:569-77. [PubMed]
- Trépo E, Nahon P, Bontempi G, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014;59:2170-7. [PubMed]
- Guyot E, Sutton A, Rufat P, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol 2013;58:312-8. [PubMed]
- Hamza S, Petit JM, Masson D, et al. PNPLA3 rs738409 GG homozygote status is associated with increased risk of heptocellular carcinoma in cirrhotic patients. J Hepatol 2012;56:S281-2.
- Nischalke HD, Berger C, Luda C, et al. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS One 2011;6:e27087. [PubMed]
- Trepo E, Guyot E, Ganne-Carrie N, et al. PNPLA3 (rs738409 C>G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology 2012;55:1307-8. [PubMed]
- Kotronen A, Johansson LE, Johansson LM, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia 2009;52:1056-60. [PubMed]
- Romeo S, Sentinelli F, Cambuli VM, et al. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol 2010;53:335-8. [PubMed]
- Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 2008;83:520-8. [PubMed]
- Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet 2010;11:172. [PubMed]
- Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 2010;51:1209-17. [PubMed]
- Burza MA, Pirazzi C, Maglio C, et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis 2012;44:1037-41. [PubMed]
- Liu YL, Patman GL, Leathart JB, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol 2014;61:75-81. [PubMed]
- Pingitore P, Pirazzi C, Mancina RM, et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta 2014;1841:574-80.
- Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem 2011;286:37085-93. [PubMed]
- Jenkins CM, Mancuso DJ, Yan W, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 2004;279:48968-75. [PubMed]
- Pirazzi C, Adiels M, Burza MA, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276-82. [PubMed]
- He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010;285:6706-15. [PubMed]
- Perttilä J, Huaman-Samanez C, Caron S, et al. PNPLA3 is regulated by glucose in human hepatocytes, and its I148M mutant slows down triglyceride hydrolysis. Am J Physiol Endocrinol Metab 2012;302:E1063-9. [PubMed]
- Li JZ, Huang Y, Karaman R, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest 2012;122:4130-44. [PubMed]
- Basantani MK, Sitnick MT, Cai L, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res 2011;52:318-29. [PubMed]
- Qiao A, Liang J, Ke Y, et al. Mouse patatin-like phospholipase domain-containing 3 influences systemic lipid and glucose homeostasis. Hepatology 2011;54:509-21. [PubMed]
- Chen W, Chang B, Li L, et al. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology 2010;52:1134-42. [PubMed]
- Smagris E. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015;61:108-18. [PubMed]
- Rydel TJ, Williams JM, Krieger E, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 2003;42:6696-708. [PubMed]
- Wilson PA, Gardner SD, Lambie NM, et al. Characterization of the human patatin-like phospholipase family. J Lipid Res 2006;47:1940-9. [PubMed]
- Huang Y, He S, Li JZ, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A 2010;107:7892-7. [PubMed]
- Ruhanen H, Perttilä J, Hölttä-Vuori M, et al. PNPLA3 mediates hepatocyte triacylglycerol remodeling. J Lipid Res 2014;55:739-46. [PubMed]
- Sevastianova K, Santos A, Kotronen A, et al. Effect of short-term carbohydrate overfeeding and long-term weight loss on liver fat in overweight humans. Am J Clin Nutr 2012;96:727-34. [PubMed]
- Havel RJ. Lipoproteins and lipid transport. Adv Exp Med Biol 1975;63:37-59. [PubMed]
- Olofsson SO, Asp L, Borén J. The assembly and secretion of apolipoprotein B-containing lipoproteins. Curr Opin Lipidol 1999;10:341-6. [PubMed]
- Sen D, Dagdelen S, Erbas T. Hepatosteatosis with hypobetalipoproteinemia. J Natl Med Assoc 2007;99:284-6. [PubMed]
- Kumashiro N, Yoshimura T, Cantley JL, et al. Role of patatin-like phospholipase domain-containing 3 on lipid-induced hepatic steatosis and insulin resistance in rats. Hepatology 2013;57:1763-72. [PubMed]
- Shimomura I, Bashmakov Y, Ikemoto S, et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci U S A 1999;96:13656-61. [PubMed]
- Palmer CN, Maglio C, Pirazzi C, et al. Paradoxical lower serum triglyceride levels and higher type 2 diabetes mellitus susceptibility in obese individuals with the PNPLA3 148M variant. PLoS One 2012;7:e39362. [PubMed]
- Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet 2014;23:4077-85. [PubMed]
- Pitt GA. Chemical structure and the changing concept of vitamin A activity. Proc Nutr Soc 1983;42:43-51. [PubMed]
- Blomhoff R, Rasmussen M, Nilsson A, et al. Hepatic retinol metabolism. Distribution of retinoids, enzymes, and binding proteins in isolated rat liver cells. J Biol Chem 1985;260:13560-5. [PubMed]
- Blomhoff R, Helgerud P, Rasmussen M, et al. In vivo uptake of chylomicron [3H]retinyl ester by rat liver: evidence for retinol transfer from parenchymal to nonparenchymal cells. Proc Natl Acad Sci U S A 1982;79:7326-30. [PubMed]
- Weber F. Biochemical mechanisms of vitamin A action. Proc Nutr Soc 1983;42:31-41. [PubMed]
- Newcomer ME, Ong DE. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim Biophys Acta 2000;1482:57-64. [PubMed]
- Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 2008;88:125-72. [PubMed]


