Landscape of genomic alterations in hepatocellular carcinoma: current knowledge and perspectives for targeted therapies
Hepatocellular carcinoma (HCC) is a deadly cancer worldwide characterized by a rising incidence and limited therapeutic options (1). HCC is associated with multiple risk factors, including chronic hepatitis B and C (HBV/HCV), alcohol intake, aflatoxin B1 exposure, diabetes and obesity. In addition, liver carcinogenesis is a long and multistep process associated with the accumulation of multiple genetic and epigenetic alterations resulting in tumor heterogeneity (1). Thus, each HCC tumor is characterized by a unique molecular fingerprint made of a specific combination of somatic alterations (2). Deciphering this molecular fingerprint is essential to develop effective personalized treatments. Over the last decades, genome-wide unsupervised strategies including next-generation sequencing (NGS) and gene expression profiling allowed a deep characterization of genomic alterations in HCC, as well as the definition of clinically relevant HCC subtypes (3-6). In direct continuity with this experienced strategy, The Cancer Genome Atlas (TCGA) network recently reported a comprehensive and integrative genomic characterization of a large cohort of 363 HCC (7). A multiplex molecular analysis was performed, including exome sequencing, DNA copy number and methylation analysis, gene expression profiling, and proteomics (Figure 1). The study validates known and identifies novel driver gene candidates in HCC, defines putative key therapeutic targets, and data integration from multiple genomic platforms provides mechanistic molecular insights for the observed alterations (7).
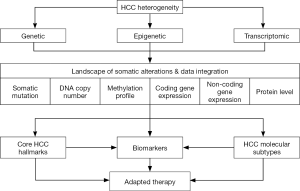
Whole-genome and exome sequencing provided an accurate landscape of recurrent somatic mutations and driver genes in HCC (3,4,7). The most frequently observed mutational events include TERT promoter activating mutations (~40% HCC), and the mutually exclusive mutations of TP53 and CTNNB1 genes (~30% HCC). Taken together, these studies highlighted a complex network of signaling pathways that constitute the core functional hallmarks of HCC (5,6,8).
Telomere maintenance
Reactivation of telomerase is an early and recurrent event in liver carcinogenesis as it is detected in preneoplastic lesions and in >90% HCC. These observations open new opportunities for early HCC detection, specifically if TERT alterations could be detected in circulating tumor cells. Sequencing analysis demonstrated that TERT reactivation mainly results from activating mutations or viral insertion at the promoter site, and focal DNA amplification (8). Interestingly, HCV-associated HCC are more likely to be mutated in TERT promoter, as compared to HBV-associated HCC in which TERT reactivation is frequently linked to HBV integration. TERT promoter mutations are also frequently associated with CTNNB1 mutations, suggesting a cooperation of the two pathways in liver tumorigenesis. Thus, interfering with telomerase and the Wnt/β-catenin pathway may represent a promising strategy.
Wnt/β-catenin pathway
The Wnt/β-catenin pathway plays a key role in liver physiology and is frequently altered in HCC. Although not addressed in the TCGA study, a specific spectrum of CTNNB1 activating mutations is closely associated with the activity of β-catenin and its oncogenic potential in HCC (9). Negative regulators of the Wnt/β-catenin pathway, including AXIN1/2 and APC, are also inactivated by mutations in HCC. Thus, various strategies have been developed to interfere with the Wnt/β-catenin pathway, e.g., by using small intracellular inhibitors or soluble decoy receptors for Wnt ligands. However, such pharmacological inhibitors have not been translated into the clinic yet, due to the difficulty of designing highly selective and non-toxic molecules (8). A better characterization of canonical and non-canonical effects of the Wnt/β-catenin pathway and its crosstalk with the tumor microenvironment may help to model potential side effects of such inhibitors and to design effective combined therapies. Notably, an association of Wnt and TGFβ inhibitors may be relevant in Hoshida’s S1 HCC subtype in which a crosstalk between Wnt/β-catenin and TGFβ pathways has been previously identified (6).
Cell cycle control
The P53 cell cycle pathway is frequently altered in HCC (8). Interestingly, by using a signature of P53 transcriptional targets, the TCGA study identifies a group of P53 inactive tumors but independently to TP53 mutations. The amplification of P53 inhibitor MDM4 is notably identified as an alternate mechanism of P53 inactivation (7). P53 inactive HCC subtype is associated with a poor prognosis, stemness features and activation of the sonic hedgehog signaling (7). DNA hypermethylation was also found to be the major mechanism of silencing of cyclin dependent kinase inhibitor CDKN2A (>50% HCC) whereas mutations of CDKN2A and other cell cycle regulators (e.g., RB1, CCNE1) occur in less than 4% HCC (7). Frequent alterations of numerous targets linked to the activity of proliferation-associated receptor tyrosine kinases (RTK) (e.g., RAS, PI3K, PTEN) are in agreement with the sensitivity of HCC to RTK inhibitors (10).
Chromatin remodeling
As previously reported, recurrent somatic mutations of chromatin modifier genes, including ARID1/2, BAP1 or histone methyl transferases KMT2C and KMT2D were identified (7). Although this point was not addressed in the TCGA study, integration of long non-coding RNA profiles would also provide mechanistic insights into HCC tumorigenesis. Indeed, long non-coding RNA play a key role in epigenetics, notably by acting as scaffolds for chromatin remodeling factors (11). Changes in chromatin remodeling or DNA methylation as a result of somatic mutations or gene expression alterations suggest that therapeutic targeting of epigenetic pathways is relevant in HCC. Accordingly, by integrating multiple transcriptomic profiles, we recently reported that histone deacetylase inhibitors may target multiple core hallmarks of HCC (5).
Molecular HCC subtyping
Molecular stratification of HCC has been extensively described based on genetic, epigenetic and transcriptomic profiles. The originality of the TCGA study comes from the integration of multiple genomic data types using a joint latent variable model (12). The so-called iCluster algorithm identified 3 HCC subtypes (iC1-3) which partially overlap with S1–3 subtypes, as defined by Hoshida (6). However, the absence of significant differences in term of survival between the 3 iClusters (except for the poor prognosis iC1 subtype) challenges the approach of integrating all genomic data types simultaneously. It will be interesting to determine whether some specific data types (e.g., quantitative mRNA levels) are more efficient than others for patient stratification.
Novel HCC driver genes
The TGCA study identified 8 novel HCC driver candidates (LZTR1, AZIN1, RP1L1, EEF1A1, GPATCH4, CREB3L3, AHCTF1, HIST1H1C) mutated in 1–4% HCC (7). Functional studies and modelling of the impact of the identified mutations will be required to validate these genes as true drivers in HCC. Interestingly, cholangiocarcinoma-associated IDH1/2 mutations were identified in 4 tumors. Although exhibiting histological features of HCC, integrating gene expression profiles of IDH1/2 mutated tumors with those of well-defined HCC, intrahepatic cholangiocarcinoma (iCCA) and mixed iCCA/HCC tumors may help to clarify their cellular origin. Indeed, these HCC were associated with features previously observed in iCCA and mixed iCCA/HCC, including a very poor prognosis and the expression of stemness markers (13,14). Thus, the existence of IDH1/2 mutated HCC supports the hypothesis of tumors possibly arising from cancer stem cells.
Immune-based therapeutic strategies
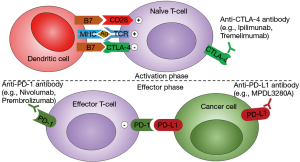
Striking differences in the relative composition of immune cell populations were observed between non-tumor and tumor tissues highlighting a switch from activating effector cells to resting suppressive immune cells. Based on the expression of 66 curated immune cell markers, a HCC subset with high expression of immune checkpoint genes (e.g., CTLA4, PDCD1/PD1, CD274/PDL1) was identified, opening interesting opportunities for efficient targeted therapies using immune checkpoint inhibitors, e.g., anti-CTLA4 and anti-PD1/PDL1 antibodies (Figure 2).
In conclusion, integrative genomic studies, including the TCGA study, clearly point out the heterogeneity of HCC alterations and suggest that combined therapies targeting several cancer hallmarks simultaneously may represent a clinically relevant strategy (Figure 1). Once a consensus in HCC subtyping will be established, another important step to be reached for a better clinical translation will be the definition of specific biomarkers for each HCC subtype, ideally detectable with poorly invasive methods. In this context, further investigations will be required to determine whether specific alterations reflecting each HCC subtype could be detected in liquid biopsies.
Acknowledgements
Funding: Author’s laboratory is supported by funds from Inserm, University of Rennes, Institut National du Cancer, Ligue Contre le Cancer (cd22, cd35, cd85), and Association Française pour l’Etude du Foie (AFEF). KB is supported by a doctoral fellowship from Ligue Contre le Cancer (cd22) and Conseil Régional de Bretagne.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet 2002;31:339-46. [Crossref] [PubMed]
- Schulze K, Nault JC, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol 2016;65:1031-42. [Crossref] [PubMed]
- Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505-11. [Crossref] [PubMed]
- Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73. [Crossref] [PubMed]
- Allain C, Angenard G, Clement B, et al. Integrative Genomic Analysis Identifies the Core Transcriptional Hallmarks of Human Hepatocellular Carcinoma. Cancer Res 2016;76:6374-81. [Crossref] [PubMed]
- Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 2009;69:7385-92. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017;169:1327-41.e23. [Crossref] [PubMed]
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226-39.e4. [Crossref] [PubMed]
- Rebouissou S, Franconi A, Calderaro J, et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology 2016;64:2047-61. [Crossref] [PubMed]
- Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015;12:408-24. [Crossref] [PubMed]
- Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452-63. [Crossref] [PubMed]
- Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 2009;25:2906-12. [Crossref] [PubMed]
- Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410-6. [Crossref] [PubMed]
- Coulouarn C, Cavard C, Rubbia-Brandt L, et al. Combined hepatocellular-cholangiocarcinomas exhibit progenitor features and activation of Wnt and TGFbeta signaling pathways. Carcinogenesis 2012;33:1791-6. [Crossref] [PubMed]


