Cell-free DNA methylation as liquid biopsy for the assessment of fibrosis in patients with nonalcoholic steatohepatitis: a gap between innovation and implementation
Nonalcoholic fatty liver disease (NAFLD) is a common chronic liver disease whose prevalence has reached global epidemic proportions, both in adults and children (1). There is consistent clinical and epidemiological evidence supporting the assertion that NAFLD may progress from a benign histological stage characterized by hepatic triglyceride accumulation, also known as simple steatosis, to a more severe histological picture characterized by liver cell injury, a mixed inflammatory lobular infiltrate, and variable fibrosis named nonalcoholic steatohepatitis (NASH) (1). Numerous factors influence the disease severity and progression, including genetic predisposition (2) and aberrant tissue-specific DNA methylation modifications that have been observed not only in the nuclear (1,3,4) but the mitochondrial genome as well (5). Presently, DNA methylation is one of the most extensively studied epigenetic modifications in eukaryotes consisting of a methyl group covalently added to a cytosine, yielding 5-methylcytosine (5mC).
In addition, robust evidence indicates that the progression of NAFLD is associated with both mitochondrial dysfunction (6) and genetic diversity in mitochondrial genes that encode for members of the oxidative phosphorylation (OXPHOS) chain (7). Furthermore, environmental factors such alcohol consumption, even in light or moderate amounts (8), as well as diet and lifestyle (9), are also implicated in the development of a more aggressive histological phenotype. Interestingly, available evidence indicates that lifestyle factors such as exercise may modify epigenetic marks (5).
The histological diagnosis of NASH is based exclusively on liver biopsy, which is currently the gold standard for determining the disease diagnosis and prognosis accurately (10). Unfortunately, this method is not only prone to patient complications, but is also expensive, while requiring extensive training and advanced skills. In the last decade, numerous imaging methods (11) as well as circulating molecular biomarkers, including epigenetic factors such as miRNAs (12), have been proposed, opened up the potential for the non-invasive diagnosis of liver fibrosis and advanced NASH. In fact, we have demonstrated that circulating miRNAs, in particular miR-122, not only mirror histological and molecular events occurring in the liver but have a reliable predictive power, allowing simple steatosis and NASH to be distinguished (12). In addition, we observed that NAFLD has a distinctive circulating miRNA profile associated with a global dysmetabolic disease-state and cardiovascular risk (12).
The evolving implementation of high-throughput OMICs profiling of biological samples—which includes genetic variation, metabolomics, transcriptomics, proteomics, metagenomics and epigenomics—has directly influenced the rate of biomarker discovery. Unfortunately, integration of this knowledge into the healthcare system is time consuming, while also facing other tremendous challenges that must be overcome.
Hardy and coworkers have recently published a proof-of-concept study involving a small sample of patients, which findings suggest that plasma DNA methylation signatures might reflect the molecular pathology associated with fibrotic liver disease (13). Specifically, the authors showed that differential DNA methylation at two CpG sites in the promoter of peroxisome proliferator-activated receptor γ (PPARγ) can be detected within the pool of cell-free DNA (cfDNA) of patients with end-stage liver disease, and may potentially allow stratifying patients with NAFLD into those with mild versus severe fibrosis (13). While interesting and promising, these findings deserve some reflections.
The role of epigenetics in the biology of NAFLD
Epigenetic mechanisms play a critical role in the reprogramming machinery of hepatocytes and other liver cells as a means of adapting to the stressful environment caused by the abnormal accumulation of fat and its consequences, including inflammation and oxidative stress (14). Robust evidence yielded by human studies has demonstrated that NAFLD severity is associated with a consistent pattern in the methylome and transcriptome of the liver tissue characterized by hypermethylation and down-regulation of genes involved in metabolic function (3,5,15) versus hypomethylation and up-regulation of genes involved in liver regeneration, tumorigenesis and tissue repair (15). This damage-associated molecular pattern is translated into a disease phenotype characterized by increased fibrogenesis and metabolic deregulation.
Tissue-specific methylation changes can be detected in circulation: the attractive concept of liquid biopsy
As mentioned earlier, the distinguishing epigenetic modifications associated with NAFLD progression are tissue-specific, as they are observed either in the liver of the affected patients or other tissues, while still having a direct effect on the liver functioning. Whether liver-related aberrant DNA methylation patterns can be directly assessed in the blood by exploring cfDNA, which would enable accurate stratification of liver fibrosis associated with NASH, remains to be demonstrated in large cohorts.
A remarkable study, as a part of which the researchers performed a genome-wide bisulfite sequencing of plasma cfDNA, yielded findings indicating that white blood cells are the predominant contributors to the circulating DNA pool, while cells derived from the liver are sufficiently represented as well (16). This finding certainly supports the potential for the use of methylation modifications in cfDNA as liquid biopsy in the diagnosis and prognosis of liver diseases, hepatocellular carcinoma in particular (16). Nevertheless, the finding suggesting that two promoter CpG sites in PPARγ become hypermethylated as fibrosis severity increases, where this observation served as a surrogate of specific changes occurring in the liver tissue (13), deserves some other considerations. First, cfDNA consists of a pool of mixture DNA released from different tissues of the body, usually as a byproduct of dead—mostly apoptotic—cells. Hence, the question of whether NASH-associated significant fibrosis is linked to significant hepatocyte cell death that in turn explains the source of cfDNA remains unanswered in the study conducted by Hardy et al. Unfortunately, the authors did not present any morphologic or circulating biomarkers of cell death—for instance, caspase-generated cytokeratin-18 fragments (CK-18)—in association with their findings. Extant literature provides some evidence supporting the notion that genetic diversity in genes encoding the ten-eleven-translocation (TET) family of proteins—which are responsible for catalyzing the conversion of 5-methylcytosine to 5-hmC—is involved in the epigenetic regulation of programmed liver-cell death in patients with NASH (4). Thus, the connection between advanced fibrosis and epigenetic modifications that eventually feed the pool of cfDNA in patients with NASH is still not fully elucidated.
Second, the selection of PPARγ as the candidate gene for exploring surrogate liver DNA methylation changes in the blood represents a significant challenge due to the high expression levels of this gene in many different tissues that are strongly associated with the biology of NAFLD and related co-morbidities. For example, under physiological conditions, PPARγ exhibits the highest expression in adipose tissue, and is lower in skeletal muscle, spleen, heart and the liver; however, PPARγ is also detectable in placenta, lung and ovary. A comprehensive list of tissues in which PPARγ is expressed, as well as the magnitude of its expression, is shown in Table 1. Interestingly, in disease states, including the metabolic syndrome and obesity—two conditions strongly associated with advanced fibrosis in NASH—PPARγ is hypermethylated in adipocytes (17). Furthermore, recent evidence indicates that transient PPARγ promoter methylation occurs after common events of ordinary life, for instance, physical exercise or caffeine consumption (18). Hence, dynamic changes associated with epigenetic modifications may specifically jeopardize the selection of PPARγ as the “candidate gene” for use as a biomarker of liver fibrosis.
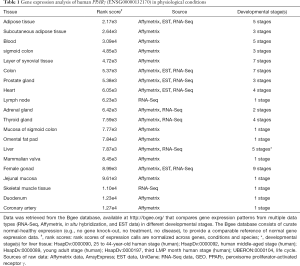
Full table
Challenges: translating laboratory work into the clinical practice
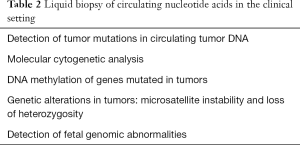
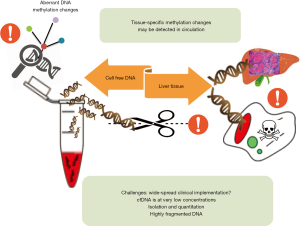
The presence of cfDNA in the circulating compartment was demonstrated three decades ago (19), leading to the emergence of liquid biopsy. The applications of this innovative concept in the clinical setting are summarized in Table 2. However, the implementation of cfDNA methylation as a non-invasive molecular tool for the diagnosis of fibrosis imposes tremendous analytical and technical challenges because cfDNA is not only highly fragmented (~170–500 bp) but also circulates at very low concentrations (1.8–44 ng/mL). The fact that exploration of DNA methylation signatures requires a previous step of DNA bisulfitation or other complex and protracted techniques, such as 5mC-containing DNA immunoprecipitation and sequencing, imposes further obstacles to the adoption of this method. The three main pitfalls to overcome before cfDNA methylation could be employed as biomarker of fibrosis are illustrated in Figure 1. Available evidence suggests that a considerable gap between innovation and implementation still exists, preventing definitive affirmation that plasma DNA methylation signatures reflect the molecular pathology associated with fibrotic liver disease. Nonetheless, this is a promising horizon toward which further research efforts should be directed.
Full table
Acknowledgements
Funding: This study was partially supported by grants PICT 2014-0432, PICT 2014-1816 and PICT 2015-0551 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT). S Sookoian and CJ Pirola belong to Consejo Nacional de Investigaciones Científicas (CONICET).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015;1:15080. [Crossref] [PubMed]
- Sookoian S, Pirola CJ. The genetic epidemiology of nonalcoholic fatty liver disease: toward a personalized medicine. Clin Liver Dis 2012;16:467-85. [Crossref] [PubMed]
- Sookoian S, Rosselli MS, Gemma C, et al. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology 2010;52:1992-2000. [Crossref] [PubMed]
- Pirola CJ, Scian R, Gianotti TF, et al. Epigenetic Modifications in the Biology of Nonalcoholic Fatty Liver Disease: The Role of DNA Hydroxymethylation and TET Proteins. Medicine (Baltimore) 2015;94:e1480. [Crossref] [PubMed]
- Pirola CJ, Gianotti TF, Burgueño AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 2013;62:1356-63. [Crossref] [PubMed]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 2001;120:1183-92. [Crossref] [PubMed]
- Sookoian S, Flichman D, Scian R, et al. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol 2016;240:437-49. [Crossref] [PubMed]
- Sookoian S, Flichman D, Castaño GO, et al. Mendelian randomisation suggests no beneficial effect of moderate alcohol consumption on the severity of nonalcoholic fatty liver disease. Aliment Pharmacol Ther 2016;44:1224-34. [Crossref] [PubMed]
- Hannah WN Jr, Harrison SA. Lifestyle and Dietary Interventions in the Management of Nonalcoholic Fatty Liver Disease. Dig Dis Sci 2016;61:1365-74. [Crossref] [PubMed]
- Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology 2012;142:1592-609. [Crossref] [PubMed]
- Hannah WN Jr, Harrison SA. Noninvasive imaging methods to determine severity of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 2016;64:2234-43. [Crossref] [PubMed]
- Pirola CJ, Fernández Gianotti T, Castaño GO, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015;64:800-12. [Crossref] [PubMed]
- Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Sookoian S, Pirola CJ. NAFLD. Metabolic make-up of NASH: from fat and sugar to amino acids. Nat Rev Gastroenterol Hepatol 2014;11:205-7. [Crossref] [PubMed]
- Murphy SK, Yang H, Moylan CA, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 2013;145:1076-87. [Crossref] [PubMed]
- Sun K, Jiang P, Chan KC, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503-12. [Crossref] [PubMed]
- Fujiki K, Kano F, Shiota K, et al. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol 2009;7:38. [Crossref] [PubMed]
- Barrès R, Yan J, Egan B, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab 2012;15:405-11. [Crossref] [PubMed]
- Shapiro B, Chakrabarty M, Cohn EM, et al. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983;51:2116-20. [Crossref] [PubMed]


